Data science in unveiling COVID-19 pathogenesis and diagnosis: evolutionary origin to drug repurposing
- PMID: 33592108
- PMCID: PMC7929414
- DOI: 10.1093/bib/bbaa420
Data science in unveiling COVID-19 pathogenesis and diagnosis: evolutionary origin to drug repurposing
Abstract
Motivation: The outbreak of novel severe acute respiratory syndrome coronavirus (SARS-CoV-2, also known as COVID-19) in Wuhan has attracted worldwide attention. SARS-CoV-2 causes severe inflammation, which can be fatal. Consequently, there has been a massive and rapid growth in research aimed at throwing light on the mechanisms of infection and the progression of the disease. With regard to this data science is playing a pivotal role in in silico analysis to gain insights into SARS-CoV-2 and the outbreak of COVID-19 in order to forecast, diagnose and come up with a drug to tackle the virus. The availability of large multiomics, radiological, bio-molecular and medical datasets requires the development of novel exploratory and predictive models, or the customisation of existing ones in order to fit the current problem. The high number of approaches generates the need for surveys to guide data scientists and medical practitioners in selecting the right tools to manage their clinical data.
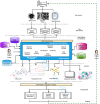
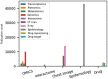
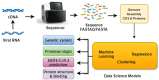
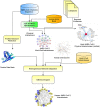
Results: Focusing on data science methodologies, we conduct a detailed study on the state-of-the-art of works tackling the current pandemic scenario. We consider various current COVID-19 data analytic domains such as phylogenetic analysis, SARS-CoV-2 genome identification, protein structure prediction, host-viral protein interactomics, clinical imaging, epidemiological research and drug discovery. We highlight data types and instances, their generation pipelines and the data science models currently in use. The current study should give a detailed sketch of the road map towards handling COVID-19 like situations by leveraging data science experts in choosing the right tools. We also summarise our review focusing on prime challenges and possible future research directions.
Contact: hguzzi@unicz.it, sroy01@cus.ac.in.
Keywords: COVID-19; SARS-CoV-2; artificial intelligence; data science; network science.
© The Author(s) 2021. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures



References
-
- Hook DW, Porter SJ, Herzog C. Dimensions: building context for search and evaluation. Front Res Metr Anal 2018;3:23.
-
- Noruzi A. Google scholar: the new generation of citation indexes. Libri 2005;55(4):170–80.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

