Identification of hub genes associated with adult acute myeloid leukemia progression through weighted gene co-expression network analysis
- PMID: 33592582
- PMCID: PMC7950274
- DOI: 10.18632/aging.202493
Identification of hub genes associated with adult acute myeloid leukemia progression through weighted gene co-expression network analysis
Abstract
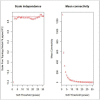
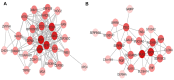
Acute myeloid leukemia (AML) is a malignancy of hematopoietic stem cells. Although many candidate genes such as CEBPA, FLT3, IDH1, and IDH2 have been associated with AML initiation and prognosis, the molecular mechanisms underlying this disease remain unclear. In this study, we used a systemic co-expression analysis method, namely weighted gene co-expression network analysis (WGCNA), to identify new candidate genes associated with adult AML progression and prognosis. We identified around 5,138 differentially expressed genes (DEGs) between AML samples (from The Cancer Genome Atlas database) and normal control samples (from the Genotype-Tissue Expression database). WGCNA identified nine co-expression modules with significant differences based on the DEGs. Among modules, the turquoise and blue ones were the most relevant to AML (P-value: turquoise 0, blue 4.64E-77). GO term and KEGG pathway analyses revealed that pathways that are commonly dysregulated in AML were all enriched in the blue and turquoise modules. A total of 15 hub genes were identified to be crucial for AML progression. PIVOT analysis revealed non-coding RNAs, transcriptional factors, and drugs associated with the hub genes. Finally, survival analysis revealed that one of the hub genes, CEACAM5, was significantly associated with AML prognosis and could serve as a potential target for AML treatment.
Keywords: AML; WGCNA; hub genes.
Conflict of interest statement
Figures




References
-
- Duployez N, Marceau-Renaut A, Villenet C, Petit A, Rousseau A, Ng SW, Paquet A, Gonzales F, Barthélémy A, Leprêtre F, Pottier N, Nelken B, Michel G, et al. The stem cell-associated gene expression signature allows risk stratification in pediatric acute myeloid leukemia. Leukemia. 2019; 33:348–57. 10.1038/s41375-018-0227-5 - DOI - PubMed
-
- Beck D, Thoms JA, Palu C, Herold T, Shah A, Olivier J, Boelen L, Huang Y, Chacon D, Brown A, Babic M, Hahn C, Perugini M, et al. A four-gene LincRNA expression signature predicts risk in multiple cohorts of acute myeloid leukemia patients. Leukemia. 2018; 32:263–72. 10.1038/leu.2017.210 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

