The effects of RNA editing in cancer tissue at different stages in carcinogenesis
- PMID: 33593231
- PMCID: PMC8582992
- DOI: 10.1080/15476286.2021.1877024
The effects of RNA editing in cancer tissue at different stages in carcinogenesis
Abstract
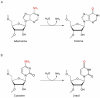
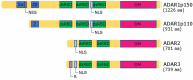
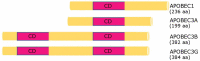
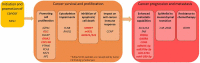
RNA editing is one of the most prevalent and abundant forms of post-transcriptional RNA modification observed in normal physiological processes and often aberrant in diseases including cancer. RNA editing changes the sequences of mRNAs, making them different from the source DNA sequence. Edited mRNAs can produce editing-recoded protein isoforms that are functionally different from the corresponding genome-encoded protein isoforms. The major type of RNA editing in mammals occurs by enzymatic deamination of adenosine to inosine (A-to-I) within double-stranded RNAs (dsRNAs) or hairpins in pre-mRNA transcripts. Enzymes that catalyse these processes belong to the adenosine deaminase acting on RNA (ADAR) family. The vast majority of knowledge on the RNA editing landscape relevant to human disease has been acquired using in vitro cancer cell culture models. The limitation of such in vitro models, however, is that the physiological or disease relevance of results obtained is not necessarily obvious. In this review we focus on discussing in vivo occurring RNA editing events that have been identified in human cancer tissue using samples surgically resected or clinically retrieved from patients. We discuss how RNA editing events occurring in tumours in vivo can identify pathological signalling mechanisms relevant to human cancer physiology which is linked to the different stages of cancer progression including initiation, promotion, survival, proliferation, immune escape and metastasis.
Keywords: ADARs; RNA editing; RNA editing in cancer; cancer development.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures
References
-
- Gallo A, Vukic D, Michalík D, et al. ADAR RNA editing in human disease; more to it than meets the I. Hum Genet. 2017;136(9):1265–1278. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
