Novel DNA methylation signatures of tobacco smoking with trans-ethnic effects
- PMID: 33593402
- PMCID: PMC7888173
- DOI: 10.1186/s13148-021-01018-4
Novel DNA methylation signatures of tobacco smoking with trans-ethnic effects
Abstract
Background: Smoking remains one of the leading preventable causes of death. Smoking leaves a strong signature on the blood methylome as shown in multiple studies using the Infinium HumanMethylation450 BeadChip. Here, we explore novel blood methylation smoking signals on the Illumina MethylationEPIC BeadChip (EPIC) array, which also targets novel CpG-sites in enhancers.
Method: A smoking-methylation meta-analysis was carried out using EPIC DNA methylation profiles in 1407 blood samples from four UK population-based cohorts, including the MRC National Survey for Health and Development (NSHD) or 1946 British birth cohort, the National Child Development Study (NCDS) or 1958 birth cohort, the 1970 British Cohort Study (BCS70), and the TwinsUK cohort (TwinsUK). The overall discovery sample included 269 current, 497 former, and 643 never smokers. Replication was pursued in 3425 trans-ethnic samples, including 2325 American Indian individuals participating in the Strong Heart Study (SHS) in 1989-1991 and 1100 African-American participants in the Genetic Epidemiology Network of Arteriopathy Study (GENOA).
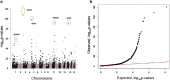
Results: Altogether 952 CpG-sites in 500 genes were differentially methylated between smokers and never smokers after Bonferroni correction. There were 526 novel smoking-associated CpG-sites only profiled by the EPIC array, of which 486 (92%) replicated in a meta-analysis of the American Indian and African-American samples. Novel CpG sites mapped both to genes containing previously identified smoking-methylation signals and to 80 novel genes not previously linked to smoking, with the strongest novel signal in SLAMF7. Comparison of former versus never smokers identified that 37 of these sites were persistently differentially methylated after cessation, where 16 represented novel signals only profiled by the EPIC array. We observed a depletion of smoking-associated signals in CpG islands and an enrichment in enhancer regions, consistent with previous results.
Conclusion: This study identified novel smoking-associated signals as possible biomarkers of exposure to smoking and may help improve our understanding of smoking-related disease risk.
Keywords: DNA methylation; Environment; Epigenetics; Lifestyle; SLAMF7; Smoking.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67(1):1–48.
-
- Bloch M, Ousingsawat J, Simon R, et al. KCNMA1 gene amplification promotes tumor cell proliferation in human prostate cancer. Oncogene. 2007;26:2525–2534. - PubMed
-
- Bollepalli S, Korhonen T, Kaprio J, Anders S, Ollikainen M. EpiSmokEr: a robust classifier to determine smoking status from DNA methylation data. Epigenomics. 2019;11(13):1469–1486. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G1001799/MRC_/Medical Research Council/United Kingdom
- R01 ES021367/ES/NIEHS NIH HHS/United States
- RC1 HL100185/HL/NHLBI NIH HHS/United States
- P42 ES010349/ES/NIEHS NIH HHS/United States
- R01 HL109319/HL/NHLBI NIH HHS/United States
- R01 ES025216/ES/NIEHS NIH HHS/United States
- MR/N01104X/1 2018-2020/WT_/Wellcome Trust/United Kingdom
- R01 HL109282/HL/NHLBI NIH HHS/United States
- R01 HL119443/HL/NHLBI NIH HHS/United States
- MR/N01104X/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_12019/2/MRC_/Medical Research Council/United Kingdom
- R01 HL109301/HL/NHLBI NIH HHS/United States
- P30 ES009089/ES/NIEHS NIH HHS/United States
- R01 HL141292/HL/NHLBI NIH HHS/United States
- G0000934/MRC_/Medical Research Council/United Kingdom
- MC_UU_00019/1/MRC_/Medical Research Council/United Kingdom
- R01 HL109315/HL/NHLBI NIH HHS/United States
- R01 HL109284/HL/NHLBI NIH HHS/United States
- U01 HL054457/HL/NHLBI NIH HHS/United States
- R01 HL090863/HL/NHLBI NIH HHS/United States
- MR/M004422/1/MRC_/Medical Research Council/United Kingdom
- 108439/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- R01 HL133221/HL/NHLBI NIH HHS/United States
- MC_UU_00011/5/MRC_/Medical Research Council/United Kingdom
- WT095219MA/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

