Protein manipulation using single copies of short peptide tags in cultured cells and in Drosophila melanogaster
- PMID: 33593816
- PMCID: PMC7990863
- DOI: 10.1242/dev.191700
Protein manipulation using single copies of short peptide tags in cultured cells and in Drosophila melanogaster
Abstract
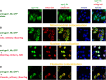
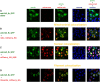
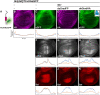
Cellular development and function rely on highly dynamic molecular interactions among proteins distributed in all cell compartments. Analysis of these interactions has been one of the main topics in cellular and developmental research, and has been mostly achieved by the manipulation of proteins of interest (POIs) at the genetic level. Although genetic strategies have significantly contributed to our current understanding, targeting specific interactions of POIs in a time- and space-controlled manner or analysing the role of POIs in dynamic cellular processes, such as cell migration or cell division, would benefit from more-direct approaches. The recent development of specific protein binders, which can be expressed and function intracellularly, along with advancement in synthetic biology, have contributed to the creation of a new toolbox for direct protein manipulations. Here, we have selected a number of short-tag epitopes for which protein binders from different scaffolds have been generated and showed that single copies of these tags allowed efficient POI binding and manipulation in living cells. Using Drosophila, we also find that single short tags can be used for POI manipulation in vivo.
Keywords: In vivo; Nanobodies; Peptide binders; Protein manipulation; Small tag.
© 2021. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures





Similar articles
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Behavioral interventions to reduce risk for sexual transmission of HIV among men who have sex with men.Cochrane Database Syst Rev. 2008 Jul 16;(3):CD001230. doi: 10.1002/14651858.CD001230.pub2. Cochrane Database Syst Rev. 2008. PMID: 18646068
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
Cited by
-
A Light-Activatable Photocaged Variant of the Ultra-High Affinity ALFA-Tag Nanobody.Chembiochem. 2022 Jun 20;23(12):e202200079. doi: 10.1002/cbic.202200079. Epub 2022 Apr 27. Chembiochem. 2022. PMID: 35411584 Free PMC article.
-
A genome-engineered tool set for Drosophila TGF-β/BMP signaling studies.Development. 2024 Nov 15;151(22):dev204222. doi: 10.1242/dev.204222. Epub 2024 Nov 18. Development. 2024. PMID: 39494616 Free PMC article.
-
Asymmetric requirement of Dpp/BMP morphogen dispersal in the Drosophila wing disc.Nat Commun. 2021 Nov 8;12(1):6435. doi: 10.1038/s41467-021-26726-6. Nat Commun. 2021. PMID: 34750371 Free PMC article.
-
Expanding the tagging toolbox for visualizing translation live.Biochem J. 2025 Jan 30;482(3):BCJ20240183. doi: 10.1042/BCJ20240183. Biochem J. 2025. PMID: 39889305 Free PMC article. Review.
-
Genetically encoded affinity reagents are a toolkit for visualizing and manipulating endogenous protein function in vivo.Nat Commun. 2025 Jul 1;16(1):5503. doi: 10.1038/s41467-025-61003-w. Nat Commun. 2025. PMID: 40595661 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

