Adult trkB Signaling in Parvalbumin Interneurons is Essential to Prefrontal Network Dynamics
- PMID: 33593856
- PMCID: PMC8026352
- DOI: 10.1523/JNEUROSCI.1848-20.2021
Adult trkB Signaling in Parvalbumin Interneurons is Essential to Prefrontal Network Dynamics
Abstract
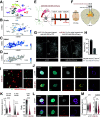
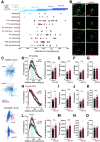
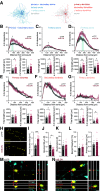
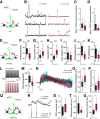
Inhibitory interneurons expressing parvalbumin (PV) are central to cortical network dynamics, generation of γ oscillations, and cognition. Dysfunction of PV interneurons disrupts cortical information processing and cognitive behavior. Brain-derived neurotrophic factor (BDNF)/tyrosine receptor kinase B (trkB) signaling regulates the maturation of cortical PV interneurons but is also implicated in their adult multidimensional functions. Using a novel viral strategy for cell-type-specific and spatially restricted expression of a dominant-negative trkB (trkB.DN), we show that BDNF/trkB signaling is essential to the integrity and maintenance of prefrontal PV interneurons in adult male and female mice. Reduced BDNF/trkB signaling in PV interneurons in the medial prefrontal cortex (mPFC) resulted in deficient PV inhibition and increased baseline local field potential (LFP) activity in a broad frequency band. The altered network activity was particularly pronounced during increased activation of the prefrontal network and was associated with changed dynamics of local excitatory neurons, as well as decreased modulation of the LFP, abnormalities that appeared to generalize across stimuli and brain states. In addition, our findings link reduced BDNF/trkB signaling in prefrontal PV interneurons to increased aggression. Together our investigations demonstrate that BDNF/trkB signaling in PV interneurons in the adult mPFC is essential to local network dynamics and cognitive behavior. Our data provide direct support for the suggested association between decreased trkB signaling, deficient PV inhibition, and altered prefrontal circuitry.SIGNIFICANCE STATEMENT Brain-derived neurotrophic factor (BDNF)/tyrosine receptor kinase B (trkB) signaling promotes the maturation of inhibitory parvalbumin (PV) interneurons, neurons central to local cortical dynamics, γ rhythms, and cognition. Here, we used a novel viral approach for reduced BDNF/trkB signaling in PV interneurons in the medial prefrontal cortex (mPFC) to establish the role of BDNF/trkB signaling in adult prefrontal network activities. Reduced BDNF/trkB signaling caused pronounced morphologic alterations, reduced PV inhibition, and deficient prefrontal network dynamics. The altered network activity appeared to manifest across stimuli and brain states and was associated with aberrant local field potential (LFP) activities and increased aggression. The results demonstrate that adult BDNF/trkB signaling is essential to PV inhibition and prefrontal circuit function and directly links BDNF/trkB signaling to network integrity in the adult brain.
Keywords: cortical inhibition; dominant-negative receptor; medial prefrontal cortex; neurotrophins; social behavior; γ oscillations.
Copyright © 2021 the authors.
Figures



Similar articles
-
δ-Containing GABAA receptors on parvalbumin interneurons modulate neuronal excitability and network dynamics in the mouse medial prefrontal cortex.J Neurophysiol. 2025 Apr 1;133(4):1003-1013. doi: 10.1152/jn.00495.2024. Epub 2025 Feb 27. J Neurophysiol. 2025. PMID: 40013665 Free PMC article.
-
Sex-specific spatial memory deficits in mice with a conditional TrkB deletion on parvalbumin interneurons.Behav Brain Res. 2019 Oct 17;372:111984. doi: 10.1016/j.bbr.2019.111984. Epub 2019 May 28. Behav Brain Res. 2019. PMID: 31150746
-
Loss of TrkB Signaling in Parvalbumin-Expressing Basket Cells Results in Network Activity Disruption and Abnormal Behavior.Cereb Cortex. 2018 Oct 1;28(10):3399-3413. doi: 10.1093/cercor/bhx173. Cereb Cortex. 2018. PMID: 28968898 Free PMC article.
-
Prefrontal and Hippocampal Parvalbumin Interneurons in Animal Models for Schizophrenia: A Systematic Review and Meta-analysis.Schizophr Bull. 2024 Jan 1;50(1):210-223. doi: 10.1093/schbul/sbad123. Schizophr Bull. 2024. PMID: 37584417 Free PMC article.
-
Differential expression of cytoskeletal regulatory factors in the adolescent prefrontal cortex: Implications for cortical development.J Neurosci Res. 2017 May;95(5):1123-1143. doi: 10.1002/jnr.23960. Epub 2016 Oct 13. J Neurosci Res. 2017. PMID: 27735056 Free PMC article. Review.
Cited by
-
Hippocampal parvalbumin interneurons play a critical role in memory development.Cell Rep. 2022 Nov 15;41(7):111643. doi: 10.1016/j.celrep.2022.111643. Cell Rep. 2022. PMID: 36384113 Free PMC article.
-
The Presynaptic Scaffold Protein Bassoon in Forebrain Excitatory Neurons Mediates Hippocampal Circuit Maturation: Potential Involvement of TrkB Signalling.Int J Mol Sci. 2021 Jul 26;22(15):7944. doi: 10.3390/ijms22157944. Int J Mol Sci. 2021. PMID: 34360710 Free PMC article.
-
Candidate Key Proteins in Tinnitus-A Bioinformatic Study of Synaptic Transmission in the Inferior Colliculus.Int J Mol Sci. 2025 Feb 20;26(5):1831. doi: 10.3390/ijms26051831. Int J Mol Sci. 2025. PMID: 40076458 Free PMC article.
-
Loss of Ezh2 in the medial ganglionic eminence alters interneuron fate, cell morphology and gene expression profiles.Front Cell Neurosci. 2024 Feb 14;18:1334244. doi: 10.3389/fncel.2024.1334244. eCollection 2024. Front Cell Neurosci. 2024. PMID: 38419656 Free PMC article.
-
Network Asynchrony Underlying Increased Broadband Gamma Power.J Neurosci. 2021 Mar 31;41(13):2944-2963. doi: 10.1523/JNEUROSCI.2250-20.2021. Epub 2021 Feb 16. J Neurosci. 2021. PMID: 33593859 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
