A Heterogeneously Expressed Gene Family Modulates the Biofilm Architecture and Hypoxic Growth of Aspergillus fumigatus
- PMID: 33593969
- PMCID: PMC8545126
- DOI: 10.1128/mBio.03579-20
A Heterogeneously Expressed Gene Family Modulates the Biofilm Architecture and Hypoxic Growth of Aspergillus fumigatus
Abstract
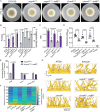
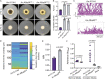
The genus Aspergillus encompasses human pathogens such as Aspergillus fumigatus and industrial powerhouses such as Aspergillus niger In both cases, Aspergillus biofilms have consequences for infection outcomes and yields of economically important products. However, the molecular components influencing filamentous fungal biofilm development, structure, and function remain ill defined. Macroscopic colony morphology is an indicator of underlying biofilm architecture and fungal physiology. A hypoxia-locked colony morphotype of A. fumigatus has abundant colony furrows that coincide with a reduction in vertically oriented hyphae within biofilms and increased low oxygen growth and virulence. Investigation of this morphotype has led to the identification of the causative gene, biofilm architecture factor A (bafA), a small cryptic open reading frame within a subtelomeric gene cluster. BafA is sufficient to induce the hypoxia-locked colony morphology and biofilm architecture in A. fumigatus Analysis across a large population of A. fumigatus isolates identified a larger family of baf genes, all of which have the capacity to modulate hyphal architecture, biofilm development, and hypoxic growth. Furthermore, introduction of A. fumigatusbafA into A. niger is sufficient to generate the hypoxia-locked colony morphology, biofilm architecture, and increased hypoxic growth. Together, these data indicate the potential broad impacts of this previously uncharacterized family of small genes to modulate biofilm architecture and function in clinical and industrial settings.IMPORTANCE The manipulation of microbial biofilms in industrial and clinical applications remains a difficult task. The problem is particularly acute with regard to filamentous fungal biofilms for which molecular mechanisms of biofilm formation, maintenance, and function are only just being elucidated. Here, we describe a family of small genes heterogeneously expressed across Aspergillus fumigatus strains that are capable of modifying colony biofilm morphology and microscopic hyphal architecture. Specifically, these genes are implicated in the formation of a hypoxia-locked colony morphotype that is associated with increased virulence of A. fumigatus Synthetic introduction of these gene family members, here referred to as biofilm architecture factors, in both A. fumigatus and A. niger additionally modulates low oxygen growth and surface adherence. Thus, these genes are candidates for genetic manipulation of biofilm development in aspergilli.
Keywords: Aspergillus fumigatus; biofilm; cryptic gene; genetics; hypoxia; morphology.
Copyright © 2021 Kowalski et al.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
