Positioning of nucleosomes containing γ-H2AX precedes active DNA demethylation and transcription initiation
- PMID: 33594057
- PMCID: PMC7886895
- DOI: 10.1038/s41467-021-21227-y
Positioning of nucleosomes containing γ-H2AX precedes active DNA demethylation and transcription initiation
Abstract
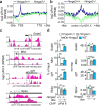
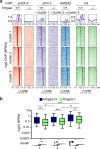
In addition to nucleosomes, chromatin contains non-histone chromatin-associated proteins, of which the high-mobility group proteins are the most abundant. Chromatin-mediated regulation of transcription involves DNA methylation and histone modifications. However, the order of events and the precise function of high-mobility group proteins during transcription initiation remain unclear. Here we show that high-mobility group AT-hook 2 protein (HMGA2) induces DNA nicks at the transcription start site, which are required by the histone chaperone FACT complex to incorporate nucleosomes containing the histone variant H2A.X. Further, phosphorylation of H2A.X at S139 (γ-H2AX) is required for repair-mediated DNA demethylation and transcription activation. The relevance of these findings is demonstrated within the context of TGFB1 signaling and idiopathic pulmonary fibrosis, suggesting therapies against this lethal disease. Our data support the concept that chromatin opening during transcriptional initiation involves intermediates with DNA breaks that subsequently require DNA repair mechanisms to ensure genome integrity.
Conflict of interest statement
G.B. was scientific advisor for a company in USA. The remaining authors declare that they have no competing interests.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

