ZMYND11-MBTD1 induces leukemogenesis through hijacking NuA4/TIP60 acetyltransferase complex and a PWWP-mediated chromatin association mechanism
- PMID: 33594072
- PMCID: PMC7886901
- DOI: 10.1038/s41467-021-21357-3
ZMYND11-MBTD1 induces leukemogenesis through hijacking NuA4/TIP60 acetyltransferase complex and a PWWP-mediated chromatin association mechanism
Abstract
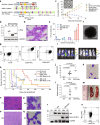
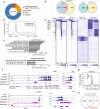
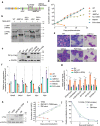
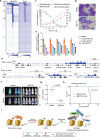
Recurring chromosomal translocation t(10;17)(p15;q21) present in a subset of human acute myeloid leukemia (AML) patients creates an aberrant fusion gene termed ZMYND11-MBTD1 (ZM); however, its function remains undetermined. Here, we show that ZM confers primary murine hematopoietic stem/progenitor cells indefinite self-renewal capability ex vivo and causes AML in vivo. Genomics profilings reveal that ZM directly binds to and maintains high expression of pro-leukemic genes including Hoxa, Meis1, Myb, Myc and Sox4. Mechanistically, ZM recruits the NuA4/Tip60 histone acetyltransferase complex to cis-regulatory elements, sustaining an active chromatin state enriched in histone acetylation and devoid of repressive histone marks. Systematic mutagenesis of ZM demonstrates essential requirements of Tip60 interaction and an H3K36me3-binding PWWP (Pro-Trp-Trp-Pro) domain for oncogenesis. Inhibitor of histone acetylation-'reading' bromodomain proteins, which act downstream of ZM, is efficacious in treating ZM-induced AML. Collectively, this study demonstrates AML-causing effects of ZM, examines its gene-regulatory roles, and reports an attractive mechanism-guided therapeutic strategy.
Conflict of interest statement
G.G.W. received research fund from the Deerfield Management/Pinnacle Hill Company. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA211336/CA/NCI NIH HHS/United States
- P30 CA016086/CA/NCI NIH HHS/United States
- R01 CA236209/CA/NCI NIH HHS/United States
- UL1 TR003107/TR/NCATS NIH HHS/United States
- TL1 TR003109/TR/NCATS NIH HHS/United States
- R01 GM122749/GM/NIGMS NIH HHS/United States
- TL1 TR002557/TR/NCATS NIH HHS/United States
- P20 GM121293/GM/NIGMS NIH HHS/United States
- R01 CA215284/CA/NCI NIH HHS/United States
- S10 OD028504/OD/NIH HHS/United States
- R01 AG024379/AG/NIA NIH HHS/United States
- S10 OD025132/OD/NIH HHS/United States
- R24 GM137786/GM/NIGMS NIH HHS/United States
- S10 OD018445/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

