This is a preprint.
Integrated plasma proteomic and single-cell immune signaling network signatures demarcate mild, moderate, and severe COVID-19
- PMID: 33594362
- PMCID: PMC7885914
- DOI: 10.1101/2021.02.09.430269
Integrated plasma proteomic and single-cell immune signaling network signatures demarcate mild, moderate, and severe COVID-19
Update in
-
Integrated plasma proteomic and single-cell immune signaling network signatures demarcate mild, moderate, and severe COVID-19.Cell Rep Med. 2022 Jul 19;3(7):100680. doi: 10.1016/j.xcrm.2022.100680. Epub 2022 Jun 28. Cell Rep Med. 2022. PMID: 35839768 Free PMC article.
Abstract
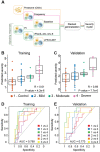
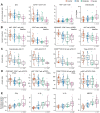
The biological determinants of the wide spectrum of COVID-19 clinical manifestations are not fully understood. Here, over 1400 plasma proteins and 2600 single-cell immune features comprising cell phenotype, basal signaling activity, and signaling responses to inflammatory ligands were assessed in peripheral blood from patients with mild, moderate, and severe COVID-19, at the time of diagnosis. Using an integrated computational approach to analyze the combined plasma and single-cell proteomic data, we identified and independently validated a multivariate model classifying COVID-19 severity (multi-class AUCtraining = 0.799, p-value = 4.2e-6; multi-class AUCvalidation = 0.773, p-value = 7.7e-6). Features of this high-dimensional model recapitulated recent COVID-19 related observations of immune perturbations, and revealed novel biological signatures of severity, including the mobilization of elements of the renin-angiotensin system and primary hemostasis, as well as dysregulation of JAK/STAT, MAPK/mTOR, and NF-κB immune signaling networks. These results provide a set of early determinants of COVID-19 severity that may point to therapeutic targets for the prevention of COVID-19 progression.
Figures


References
-
- Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R., Scott M., Hagan T., Sigal N., Feng Y., Bristow L., Tak-Yin Tsang O., Wagh D., Coller J., Pellegrini K.L., Kazmin D., Alaaeddine G., Leung W.S., Chan J.M.C., Chik T.S.H., Choi C.Y.C., Huerta C., Paine McCullough M., Lv H., Anderson E., Edupuganti S., Upadhyay A.A., Bosinger S.E., Maecker H.T., Khatri P., Rouphael N., Peiris M., and Pulendran B.. 2020. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 369:1210–1220. - PMC - PubMed
-
- Assarsson E., Lundberg M., Holmquist G., Bjorkesten J., Thorsen S.B., Ekman D., Eriksson A., Rennel Dickens E., Ohlsson S., Edfeldt G., Andersson A.C., Lindstedt P., Stenvang J., Gullberg M., and Fredriksson S.. 2014. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One 9:e95192. - PMC - PubMed
-
- Bach F.R. 2008. Bolasso: model consistent Lasso estimation through the bootstrap. ICML ‘08: Proceedings of the 25th international conference on Machine learning 33–40.
-
- Benjamini B., and Hochberg Y.. 1995. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B (Methodological) 57:289–300.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
