This is a preprint.
Disruption of nuclear architecture as a cause of COVID-19 induced anosmia
- PMID: 33594368
- PMCID: PMC7885920
- DOI: 10.1101/2021.02.09.430314
Disruption of nuclear architecture as a cause of COVID-19 induced anosmia
Update in
-
Non-cell-autonomous disruption of nuclear architecture as a potential cause of COVID-19-induced anosmia.Cell. 2022 Mar 17;185(6):1052-1064.e12. doi: 10.1016/j.cell.2022.01.024. Epub 2022 Feb 2. Cell. 2022. PMID: 35180380 Free PMC article.
Abstract
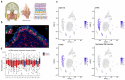
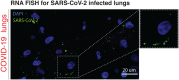
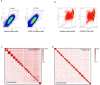
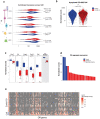
Olfaction relies on a coordinated partnership between odorant flow and neuronal communication. Disruption in our ability to detect odors, or anosmia, has emerged as a hallmark symptom of infection with SARS-CoV-2, yet the mechanism behind this abrupt sensory deficit remains elusive. Here, using molecular evaluation of human olfactory epithelium (OE) from subjects succumbing to COVID-19 and a hamster model of SARS-CoV-2 infection, we discovered widespread downregulation of olfactory receptors (ORs) as well as key components of their signaling pathway. OR downregulation likely represents a non-cell autonomous effect, since SARS-CoV-2 detection in OSNs is extremely rare both in human and hamster OEs. A likely explanation for the reduction of OR transcription is the striking reorganization of nuclear architecture observed in the OSN lineage, which disrupts multi-chromosomal compartments regulating OR expression in humans and hamsters. Our experiments uncover a novel molecular mechanism by which a virus with a very selective tropism can elicit persistent transcriptional changes in cells that evade it, contributing to the severity of COVID-19.
Figures






References
-
- Tong J. Y., Wong A., Zhu D., Fastenberg J. H. & Tham T. The Prevalence of Olfactory and Gustatory Dysfunction in COVID-19 Patients: A Systematic Review and Meta-analysis. Otolaryngology--head and neck surgery : official journal of American Academy of Otolaryngology-Head and Neck Surgery, 194599820926473, doi: 10.1177/0194599820926473 (2020). - DOI - PubMed
-
- Gerkin R. C. et al. Recent smell loss is the best predictor of COVID-19: a preregistered, cross-sectional study. medRxiv, doi: 10.1101/2020.07.22.20157263 (2020). - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
