This is a preprint.
Early introductions and community transmission of SARS-CoV-2 variant B.1.1.7 in the United States
- PMID: 33594373
- PMCID: PMC7885932
- DOI: 10.1101/2021.02.10.21251540
Early introductions and community transmission of SARS-CoV-2 variant B.1.1.7 in the United States
Update in
-
Early introductions and transmission of SARS-CoV-2 variant B.1.1.7 in the United States.Cell. 2021 May 13;184(10):2595-2604.e13. doi: 10.1016/j.cell.2021.03.061. Epub 2021 Apr 3. Cell. 2021. PMID: 33891875 Free PMC article.
Abstract
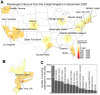
The emergence and spread of SARS-CoV-2 lineage B.1.1.7, first detected in the United Kingdom, has become a global public health concern because of its increased transmissibility. Over 2500 COVID-19 cases associated with this variant have been detected in the US since December 2020, but the extent of establishment is relatively unknown. Using travel, genomic, and diagnostic data, we highlight the primary ports of entry for B.1.1.7 in the US and locations of possible underreporting of B.1.1.7 cases. Furthermore, we found evidence for many independent B.1.1.7 establishments starting in early December 2020, followed by interstate spread by the end of the month. Finally, we project that B.1.1.7 will be the dominant lineage in many states by mid to late March. Thus, genomic surveillance for B.1.1.7 and other variants urgently needs to be enhanced to better inform the public health response.
Conflict of interest statement
Declarations of Interests M.J.M, G.K., J.M., J.T.D., M.N., N.B., and C.E.M. work for Tempus Labs. K.S.G. receives research support from Thermo Fisher for the development of assays for the detection and characterization of viruses. All other authors declare no competing interests.
Figures



References
-
- Becker R.A., Wilks A.R., Brownrigg R., Minka T.P., and Deckmyn A. (2018). maps: Draw Geographical Maps. R package version 3.3. 0.
-
- CDC (2021a). US COVID-19 Cases Caused by Variants.
-
- CDC (2021b). Genomic Surveillance for SARS-CoV-2.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
