The Role of Cytoplasmic MEX-5/6 Polarity in Asymmetric Cell Division
- PMID: 33594535
- PMCID: PMC7886744
- DOI: 10.1007/s11538-021-00860-0
The Role of Cytoplasmic MEX-5/6 Polarity in Asymmetric Cell Division
Abstract
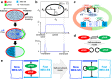
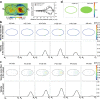
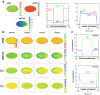
In the process of asymmetric cell division, the mother cell induces polarity in both the membrane and the cytosol by distributing substrates and components asymmetrically. Such polarity formation results from the harmonization of the upstream and downstream polarities between the cell membrane and the cytosol. MEX-5/6 is a well-investigated downstream cytoplasmic protein, which is deeply involved in the membrane polarity of the upstream transmembrane protein PAR in the Caenorhabditis elegans embryo. In contrast to the extensive exploration of membrane PAR polarity, cytoplasmic polarity is poorly understood, and the precise contribution of cytoplasmic polarity to the membrane PAR polarity remains largely unknown. In this study, we explored the interplay between the cytoplasmic MEX-5/6 polarity and the membrane PAR polarity by developing a mathematical model that integrates the dynamics of PAR and MEX-5/6 and reflects the cell geometry. Our investigations show that the downstream cytoplasmic protein MEX-5/6 plays an indispensable role in causing a robust upstream PAR polarity, and the integrated understanding of their interplay, including the effect of the cell geometry, is essential for the study of polarity formation in asymmetric cell division.
Keywords: Cell polarity; Pattern formation.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

