Severe acute respiratory syndrome-coronavirus-2 spike (S) protein based vaccine candidates: State of the art and future prospects
- PMID: 33594794
- PMCID: PMC7646037
- DOI: 10.1002/rmv.2183
Severe acute respiratory syndrome-coronavirus-2 spike (S) protein based vaccine candidates: State of the art and future prospects
Abstract
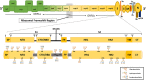
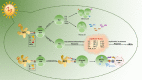
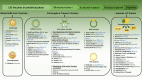
Coronavirus disease 2019 (Covid-19) is caused by severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) which is responsible for a global pandemic that started in late 2019 in Wuhan, China. To prevent the worldwide spread of this highly pathogenic virus, development of an effective and safe vaccine is urgently needed. The SARS-CoV-2 and SARS-CoV share a high degree of genetic and pathologic identity and share safety and immune-enhancement concerns regarding vaccine development. Prior animal studies with first generation (whole virus-based) preparations of SARS-CoV vaccines (inactivated and attenuated vaccine modalities) indicated the possibility of increased infectivity or eosinophilic infiltration by immunization. Therefore, development of second and third generation safer vaccines (by using modern vaccine platforms) is actively sought for this viral infection. The spike (S) protein of SARS-CoVs is the main determinant of cell entry and tropism and is responsible for facilitating zoonosis into humans and sustained person-to-person transmission. Furthermore, 'S' protein contains multiple neutralizing epitopes that play an essential role in the induction of neutralizing antibodies (nAbs) and protective immunity. Moreover, T-cell responses against the SARS-CoV-2 'S' protein have also been characterized that correlate to the IgG and IgA antibody titres in Covid-19 patients. Thus, S protein is an obvious candidate antigen for inclusion into vaccine platforms against SARS-CoV-2 viral infection. This manuscript reviews different characteristics of S protein, its potency and 'state of the art' of the vaccine development strategies and platforms using this antigen, for construction of a safe and effective SARS-CoV-2 vaccine.
Keywords: RBD; SARS-CoV-2; spike; vaccine.
© 2020 John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicting financial or other interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

