Genetic variation analyses indicate conserved SARS-CoV-2-host interaction and varied genetic adaptation in immune response factors in modern human evolution
- PMID: 33595856
- PMCID: PMC8013644
- DOI: 10.1111/dgd.12717
Genetic variation analyses indicate conserved SARS-CoV-2-host interaction and varied genetic adaptation in immune response factors in modern human evolution
Abstract
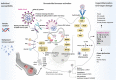
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is a pandemic as of early 2020. Upon infection, SARS-CoV-2 attaches to its receptor, that is, angiotensin-converting enzyme 2 (ACE2), on the surface of host cells and is then internalized into host cells via enzymatic machineries. This subsequently stimulates immune response factors. Since the host immune response and severity of COVID-19 vary among individuals, genetic risk factors for severe COVID-19 cases have been investigated. Our research group recently conducted a survey of genetic variants among SARS-CoV-2-interacting molecules across populations, noting near absence of difference in allele frequency spectrum between populations in these genes. Recent genome-wide association studies have identified genetic risk factors for severe COVID-19 cases in a segment of chromosome 3 that involves six genes encoding three immune-regulatory chemokine receptors and another three molecules. The risk haplotype seemed to be inherited from Neanderthals, suggesting genetic adaptation against pathogens in modern human evolution. Therefore, SARS-CoV-2 uses highly conserved molecules as its virion interaction, whereas its immune response appears to be genetically biased in individuals to some extent. We herein review the molecular process of SARS-CoV-2 infection as well as our further survey of genetic variants of its related immune effectors. We also discuss aspects of modern human evolution.
Keywords: COVID-19; genetic variant; human evolution.
© 2021 Japanese Society of Developmental Biologists.
Conflict of interest statement
On behalf of all authors, the corresponding author states that there are no conflicts of interest.
Figures
Similar articles
-
Distinct evolutionary trajectories of SARS-CoV-2-interacting proteins in bats and primates identify important host determinants of COVID-19.Proc Natl Acad Sci U S A. 2022 Aug 30;119(35):e2206610119. doi: 10.1073/pnas.2206610119. Epub 2022 Aug 10. Proc Natl Acad Sci U S A. 2022. PMID: 35947637 Free PMC article.
-
Implications of the Immune Polymorphisms of the Host and the Genetic Variability of SARS-CoV-2 in the Development of COVID-19.Viruses. 2022 Jan 6;14(1):94. doi: 10.3390/v14010094. Viruses. 2022. PMID: 35062298 Free PMC article. Review.
-
Impact of Genetic Variability in ACE2 Expression on the Evolutionary Dynamics of SARS-CoV-2 Spike D614G Mutation.Genes (Basel). 2020 Dec 24;12(1):16. doi: 10.3390/genes12010016. Genes (Basel). 2020. PMID: 33374416 Free PMC article.
-
Evaluating angiotensin-converting enzyme 2-mediated SARS-CoV-2 entry across species.J Biol Chem. 2021 Jan-Jun;296:100435. doi: 10.1016/j.jbc.2021.100435. Epub 2021 Feb 19. J Biol Chem. 2021. PMID: 33610551 Free PMC article.
-
The role of microRNAs in modulating SARS-CoV-2 infection in human cells: a systematic review.Infect Genet Evol. 2021 Jul;91:104832. doi: 10.1016/j.meegid.2021.104832. Epub 2021 Apr 1. Infect Genet Evol. 2021. PMID: 33812037 Free PMC article.
Cited by
-
Characterization of host substrates of SARS-CoV-2 main protease.Front Microbiol. 2023 Aug 21;14:1251705. doi: 10.3389/fmicb.2023.1251705. eCollection 2023. Front Microbiol. 2023. PMID: 37670988 Free PMC article. Review.
-
GWAS-significant loci and severe COVID-19: analysis of associations, link with thromboinflammation syndrome, gene-gene, and gene-environmental interactions.Front Genet. 2024 Aug 8;15:1434681. doi: 10.3389/fgene.2024.1434681. eCollection 2024. Front Genet. 2024. PMID: 39175753 Free PMC article.
-
Hospital Admission Factors Independently Affecting the Risk of Mortality of COVID-19 Patients.J Clin Med. 2023 Sep 28;12(19):6264. doi: 10.3390/jcm12196264. J Clin Med. 2023. PMID: 37834907 Free PMC article.
-
Relationships Between Polymorphisms in HLA-G 3'UTR Region and COVID-19 Disease Severity.Biochem Genet. 2024 Nov 4. doi: 10.1007/s10528-024-10951-x. Online ahead of print. Biochem Genet. 2024. PMID: 39495390
-
The etiology of preeclampsia.Am J Obstet Gynecol. 2022 Feb;226(2S):S844-S866. doi: 10.1016/j.ajog.2021.11.1356. Am J Obstet Gynecol. 2022. PMID: 35177222 Free PMC article. Review.
References
-
- Afar, D. E. , Vivanco, I. , Hubert, R. S. , Kuo, J. , Chen, E. , Saffran, D. C. , Raitano, A. B. , Jakobovits, A. (2001). Catalytic cleavage of the androgen‐regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res, 61, 1686–1692. - PubMed
Publication types
MeSH terms
Grants and funding
- U01 TR002623/TR/NCATS NIH HHS/United States
- R24OD024622/National Institution of Health
- R01MH107205/National Institution of Health
- R01 MH107205/MH/NIMH NIH HHS/United States
- 18H02983/Grant-in-Aids for Scientific Research from the Japan Society for the Promotion of Science (JSPS KAKENHI)
- PK43200005/Takeda Science Foundation
- 18K19649/Grant-in-Aids for Scientific Research from the Japan Society for the Promotion of Science (JSPS KAKENHI)
- U01TR002623/National Institution of Health
- 19K10044/Grant-in-Aids for Scientific Research from the Japan Society for the Promotion of Science (JSPS KAKENHI)
- R24 OD024622/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

