16S rRNA gene sequencing of rectal swab in patients affected by COVID-19
- PMID: 33596245
- PMCID: PMC7888592
- DOI: 10.1371/journal.pone.0247041
16S rRNA gene sequencing of rectal swab in patients affected by COVID-19
Abstract
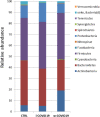
COronaVIrus Disease-2019 (COVID-19) is a pandemic respiratory infection caused by a new betacoronavirus, the Severe Acute Respiratory Syndrome-CoronaVirus-2 (SARS-CoV-2). Few data are reported on the gut microbiota in COVID-19 patients. 16S rRNA gene sequencing was performed to reveal an altered composition of the gut microbiota in patients with COVID-19 pneumonia admitted in intensive care unit (ICU) (i-COVID19), or in infectious disease wards (w-COVID19) as compared to controls (CTRL). i-COVID19 patients showed a decrease of Chao1 index as compared to CTRL and w-COVID19 patients indicating that patients in ICU displayed a lower microbial richness while no change was observed as for Shannon Index. At the phylum level, an increase of Proteobacteria was detected in w-COVID19 patients as compared to CTRL. A decrease of Fusobacteria and Spirochetes has been found, with the latter decreased in i-COVID19 patients as compared to CTRL. Significant changes in gut microbial communities in patients with COVID-19 pneumonia with different disease severity compared to CTRL have been identified. Our preliminary data may provide valuable information and promising biomarkers for the diagnosis of the disease and, when validated in larger cohort, it could facilitate the stratification of patients based on the microbial signature.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

