Ephrin receptor A2, the epithelial receptor for Epstein-Barr virus entry, is not available for efficient infection in human gastric organoids
- PMID: 33596248
- PMCID: PMC7935236
- DOI: 10.1371/journal.ppat.1009210
Ephrin receptor A2, the epithelial receptor for Epstein-Barr virus entry, is not available for efficient infection in human gastric organoids
Abstract
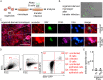
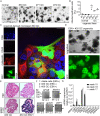
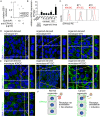
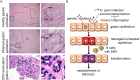
Epstein-Barr virus (EBV) is best known for infection of B cells, in which it usually establishes an asymptomatic lifelong infection, but is also associated with the development of multiple B cell lymphomas. EBV also infects epithelial cells and is associated with all cases of undifferentiated nasopharyngeal carcinoma (NPC). EBV is etiologically linked with at least 8% of gastric cancer (EBVaGC) that comprises a genetically and epigenetically distinct subset of GC. Although we have a very good understanding of B cell entry and lymphomagenesis, the sequence of events leading to EBVaGC remains poorly understood. Recently, ephrin receptor A2 (EPHA2) was proposed as the epithelial cell receptor on human cancer cell lines. Although we confirm some of these results, we demonstrate that EBV does not infect healthy adult stem cell-derived gastric organoids. In matched pairs of normal and cancer-derived organoids from the same patient, EBV only reproducibly infected the cancer organoids. While there was no clear pattern of differential expression between normal and cancer organoids for EPHA2 at the RNA and protein level, the subcellular location of the protein differed markedly. Confocal microscopy showed EPHA2 localization at the cell-cell junctions in primary cells, but not in cancer cell lines. Furthermore, histologic analysis of patient tissue revealed the absence of EBV in healthy epithelium and presence of EBV in epithelial cells from inflamed tissue. These data suggest that the EPHA2 receptor is not accessible to EBV on healthy gastric epithelial cells with intact cell-cell contacts, but either this or another, yet to be identified receptor may become accessible following cellular changes induced by inflammation or transformation, rendering changes in the cellular architecture an essential prerequisite to EBV infection.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

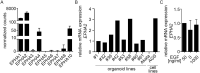
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

