Marek's disease virus US3 protein kinase phosphorylates chicken HDAC 1 and 2 and regulates viral replication and pathogenesis
- PMID: 33596269
- PMCID: PMC7920345
- DOI: 10.1371/journal.ppat.1009307
Marek's disease virus US3 protein kinase phosphorylates chicken HDAC 1 and 2 and regulates viral replication and pathogenesis
Abstract
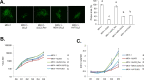
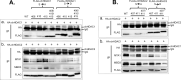
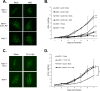
Marek's disease virus (MDV) is a potent oncogenic alphaherpesvirus that elicits a rapid onset of malignant T-cell lymphomas in chickens. Three MDV types, including GaHV-2 (MDV-1), GaHV-3 (MDV-2) and MeHV-1 (HVT), have been identified and all encode a US3 protein kinase. MDV-1 US3 is important for efficient virus growth in vitro. To study the role of US3 in MDV replication and pathogenicity, we generated an MDV-1 US3-null virus and chimeric viruses by replacing MDV-1 US3 with MDV-2 or HVT US3. Using MD as a natural virus-host model, we showed that both MDV-2 and HVT US3 partially rescued the growth deficiency of MDV-1 US3-null virus. In addition, deletion of MDV-1 US3 attenuated the virus resulting in higher survival rate and lower MDV specific tumor incidence, which could be partially compensated by MDV-2 and HVT US3. We also identified chicken histone deacetylase 1 (chHDAC1) as a common US3 substrate for all three MDV types while only US3 of MDV-1 and MDV-2 phosphorylate chHDAC2. We further determined that US3 of MDV-1 and HVT phosphorylate chHDAC1 at serine 406 (S406), while MDV-2 US3 phosphorylates S406, S410, and S415. In addition, MDV-1 US3 phosphorylates chHDAC2 at S407, while MDV-2 US3 targets S407 and S411. Furthermore, biochemical studies show that MDV US3 mediated phosphorylation of chHDAC1 and 2 affect their stability, transcriptional regulation activity, and interaction network. Using a class I HDAC specific inhibitor, we showed that MDV US3 mediated phosphorylation of chHDAC1 and 2 is involved in regulation of virus replication. Overall, we identified novel substrates for MDV US3 and characterized the role of MDV US3 in MDV pathogenesis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Schumacher D, Tischer BK, Trapp S, Osterrieder N. The protein encoded by the US3 orthologue of Marek’s disease virus is required for efficient de-envelopment of perinuclear virions and involved in actin stress fiber breakdown. J Virol. 2005;79(7):3987–97. 10.1128/JVI.79.7.3987-3997.2005 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

