Mitochondrial morphodynamics alteration induced by influenza virus infection as a new antiviral strategy
- PMID: 33596274
- PMCID: PMC7920353
- DOI: 10.1371/journal.ppat.1009340
Mitochondrial morphodynamics alteration induced by influenza virus infection as a new antiviral strategy
Erratum in
-
Correction: Mitochondrial morphodynamics alteration induced by influenza virus infection as a new antiviral strategy.PLoS Pathog. 2021 Mar 31;17(3):e1009485. doi: 10.1371/journal.ppat.1009485. eCollection 2021 Mar. PLoS Pathog. 2021. PMID: 33788903 Free PMC article.
Abstract
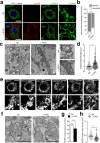
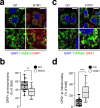
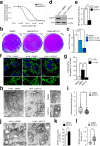
Influenza virus infections are major public health threats due to their high rates of morbidity and mortality. Upon influenza virus entry, host cells experience modifications of endomembranes, including those used for virus trafficking and replication. Here we report that influenza virus infection modifies mitochondrial morphodynamics by promoting mitochondria elongation and altering endoplasmic reticulum-mitochondria tethering in host cells. Expression of the viral RNA recapitulates these modifications inside cells. Virus induced mitochondria hyper-elongation was promoted by fission associated protein DRP1 relocalization to the cytosol, enhancing a pro-fusion status. We show that altering mitochondrial hyper-fusion with Mito-C, a novel pro-fission compound, not only restores mitochondrial morphodynamics and endoplasmic reticulum-mitochondria contact sites but also dramatically reduces influenza replication. Finally, we demonstrate that the observed Mito-C antiviral property is directly connected with the innate immunity signaling RIG-I complex at mitochondria. Our data highlight the importance of a functional interchange between mitochondrial morphodynamics and innate immunity machineries in the context of influenza viral infection.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

