The histone demethylase KDM5 is required for synaptic structure and function at the Drosophila neuromuscular junction
- PMID: 33596422
- PMCID: PMC7945993
- DOI: 10.1016/j.celrep.2021.108753
The histone demethylase KDM5 is required for synaptic structure and function at the Drosophila neuromuscular junction
Abstract
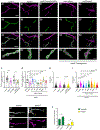
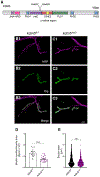
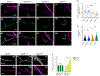
Mutations in the genes encoding the lysine demethylase 5 (KDM5) family of histone demethylases are observed in individuals with intellectual disability (ID). Despite clear evidence linking KDM5 function to neurodevelopmental pathways, how this family of proteins impacts transcriptional programs to mediate synaptic structure and activity remains unclear. Using the Drosophila larval neuromuscular junction (NMJ), we show that KDM5 is required presynaptically for neuroanatomical development and synaptic function. The Jumonji C (JmjC) domain-encoded histone demethylase activity of KDM5, which is expected to be diminished by many ID-associated alleles, is required for appropriate synaptic morphology and neurotransmission. The activity of the C5HC2 zinc finger is also required, as an ID-associated mutation in this motif reduces NMJ bouton number, increases bouton size, and alters microtubule dynamics. KDM5 therefore uses demethylase-dependent and independent mechanisms to regulate NMJ structure and activity, highlighting the complex nature by which this chromatin modifier carries out its neuronal gene-regulatory programs.
Keywords: Drosophila; KDM5; glutamatergic signaling; histone demethylase; intellectual disability; microtubule dynamics; motor neuron; neuromuscular junction; transcription.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

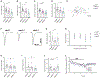
References
-
- Banovic D, Khorramshahi O, Owald D, Wichmann C, Riedt T, Fouquet W, Tian R, Sigrist SJ, and Aberle H (2010). Drosophila neuroligin 1 promotes growth and postsynaptic differentiation at glutamatergic neuromuscular junctions. Neuron 66, 724–738. - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, and Zhao K (2007). High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

