Genome-wide identification and gene-editing of pigment transporter genes in the swallowtail butterfly Papilio xuthus
- PMID: 33596834
- PMCID: PMC7891156
- DOI: 10.1186/s12864-021-07400-z
Genome-wide identification and gene-editing of pigment transporter genes in the swallowtail butterfly Papilio xuthus
Abstract
Background: Insect body coloration often functions as camouflage to survive from predators or mate selection. Transportation of pigment precursors or related metabolites from cytoplasm to subcellular pigment granules is one of the key steps in insect pigmentation and usually executed via such transporter proteins as the ATP-binding cassette (ABC) transmembrane transporters and small G-proteins (e.g. Rab protein). However, little is known about the copy numbers of pigment transporter genes in the butterfly genomes and about the roles of pigment transporters in the development of swallowtail butterflies.
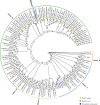
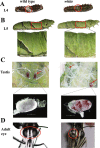
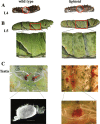
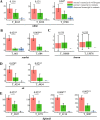
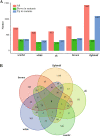
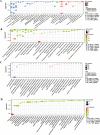
Results: Here, we have identified 56 ABC transporters and 58 Rab members in the genome of swallowtail butterfly Papilio xuthus. This is the first case of genome-wide gene copy number identification of ABC transporters in swallowtail butterflies and Rab family in lepidopteran insects. Aiming to investigate the contribution of the five genes which are orthologous to well-studied pigment transporters (ABCG: white, scarlet, brown and ok; Rab: lightoid) of fruit fly or silkworm during the development of swallowtail butterflies, we performed CRISPR/Cas9 gene-editing of these genes using P. xuthus as a model and sequenced the transcriptomes of their morphological mutants. Our results indicate that the disruption of each gene produced mutated phenotypes in the colors of larvae (cuticle, testis) and/or adult eyes in G0 individuals but have no effect on wing color. The transcriptomic data demonstrated that mutations induced by CRISPR/Cas9 can lead to the accumulation of abnormal transcripts and the decrease or dosage compensation of normal transcripts at gene expression level. Comparative transcriptomes revealed 606 ~ 772 differentially expressed genes (DEGs) in the mutants of four ABCG transporters and 1443 DEGs in the mutants of lightoid. GO and KEGG enrichment analysis showed that DEGs in ABCG transporter mutants enriched to the oxidoreductase activity, heme binding, iron ion binding process possibly related to the color display, and DEGs in lightoid mutants are enriched in glycoprotein binding and protein kinases.
Conclusions: Our data indicated these transporter proteins play an important role in body color of P. xuthus. Our study provides new insights into the function of ABC transporters and small G-proteins in the morphological development of butterflies.
Keywords: ATP-binding cassette (ABC) transporters; CRISPR/Cas9; Papilio xuthus; Rab transporters; Transcriptome.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Comprehensive microarray-based analysis for stage-specific larval camouflage pattern-associated genes in the swallowtail butterfly, Papilio xuthus.BMC Biol. 2012 May 31;10:46. doi: 10.1186/1741-7007-10-46. BMC Biol. 2012. PMID: 22651552 Free PMC article.
-
Integrated Analysis of Transcriptome and Proteome to Reveal Pupal Color Switch in Papilio xuthus Butterflies.Front Genet. 2022 Feb 3;12:795115. doi: 10.3389/fgene.2021.795115. eCollection 2021. Front Genet. 2022. PMID: 35186009 Free PMC article.
-
Species-specific coordinated gene expression and trans-regulation of larval color pattern in three swallowtail butterflies.Evol Dev. 2010 May-Jun;12(3):305-14. doi: 10.1111/j.1525-142X.2010.00416.x. Evol Dev. 2010. PMID: 20565541
-
The eyes and vision of butterflies.J Physiol. 2017 Aug 15;595(16):5457-5464. doi: 10.1113/JP273917. Epub 2017 May 8. J Physiol. 2017. PMID: 28332207 Free PMC article. Review.
-
Color and polarization vision in foraging Papilio.J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2014 Jun;200(6):513-26. doi: 10.1007/s00359-014-0903-5. Epub 2014 Apr 11. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2014. PMID: 24722674 Review.
Cited by
-
Mapping and CRISPR homology-directed repair of a recessive white eye mutation in Plodia moths.iScience. 2022 Feb 5;25(3):103885. doi: 10.1016/j.isci.2022.103885. eCollection 2022 Mar 18. iScience. 2022. PMID: 35243245 Free PMC article.
-
Developmental Transcriptome Analysis of Red-Spotted Apollo Butterfly, Parnassius bremeri.Int J Mol Sci. 2022 Sep 29;23(19):11533. doi: 10.3390/ijms231911533. Int J Mol Sci. 2022. PMID: 36232838 Free PMC article.
-
Evolutionary patterns and functional effects of 3D chromatin structures in butterflies with extensive genome rearrangements.Nat Commun. 2024 Jul 26;15(1):6303. doi: 10.1038/s41467-024-50529-0. Nat Commun. 2024. PMID: 39060230 Free PMC article.
-
Testing for fitness epistasis in a transplant experiment identifies a candidate adaptive locus in Timema stick insects.Philos Trans R Soc Lond B Biol Sci. 2022 Jul 18;377(1855):20200508. doi: 10.1098/rstb.2020.0508. Epub 2022 May 30. Philos Trans R Soc Lond B Biol Sci. 2022. PMID: 35634927 Free PMC article.
References
-
- Wang L, Kiuchi T, Fujii T, Daimon T, Li M, Banno Y, Kikuta S, Kikawada T, Katsuma S, Shimada T. Mutation of a novel ABC transporter gene is responsible for the failure to incorporate uric acid in the epidermis of ok mutants of the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2013;43(7):562–571. doi: 10.1016/j.ibmb.2013.03.011. - DOI - PubMed
-
- True JR. Insect melanism: the molecules matter. Trends Ecol Evol. 2003;18(12):640–647. doi: 10.1016/j.tree.2003.09.006. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

