Genetic diversity and population structure of early and extra-early maturing maize germplasm adapted to sub-Saharan Africa
- PMID: 33596835
- PMCID: PMC7888073
- DOI: 10.1186/s12870-021-02829-6
Genetic diversity and population structure of early and extra-early maturing maize germplasm adapted to sub-Saharan Africa
Abstract
Background: Assessment and effective utilization of genetic diversity in breeding programs is crucial for sustainable genetic improvement and rapid adaptation to changing breeding objectives. During the past two decades, the commercialization of the early and extra-early maturing cultivars has contributed to rapid expansion of maize into different agro-ecologies of sub-Saharan Africa (SSA) where maize has become an important component of the agricultural economy and played a vital role in food and nutritional security. The present study aimed at understanding the population structure and genetic variability among 439 early and extra-early maize inbred lines developed from three narrow-based and twenty-seven broad-based populations by the International Iinstitute of Tropical Agriculture Maize Improvement Program (IITA-MIP). These inbreds were genotyped using 9642 DArTseq-based single nucleotide polymorphism (SNP) markers distributed uniformly throughout the maize genome.
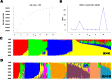
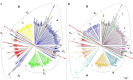
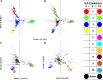
Results: About 40.8% SNP markers were found highly informative and exhibited polymorphic information content (PIC) greater than 0.25. The minor allele frequency and PIC ranged from 0.015 to 0.500 and 0.029 to 0.375, respectively. The STRUCTURE, neighbour-joining phylogenetic tree and principal coordinate analysis (PCoA) grouped the inbred lines into four major classes generally consistent with the selection history, ancestry and kernel colour of the inbreds but indicated a complex pattern of the genetic structure. The pattern of grouping of the lines based on the STRUCTURE analysis was in concordance with the results of the PCoA and suggested greater number of sub-populations (K = 10). Generally, the classification of the inbred lines into heterotic groups based on SNP markers was reasonably reliable and in agreement with defined heterotic groups of previously identified testers based on combining ability studies.
Conclusions: Complete understanding of potential heterotic groups would be difficult to portray by depending solely on molecular markers. Therefore, planned crosses involving representative testers from opposing heterotic groups would be required to refine the existing heterotic groups. It is anticipated that the present set of inbreds could contribute new beneficial alleles for population improvement, development of hybrids and lines with potential to strengthen future breeding programs. Results of this study would help breeders in formulating breeding strategies for genetic enhancement and sustainable maize production in SSA.
Keywords: DArT markers; Genetic diversity; Heterotic grouping; Maize (Zea mays L.); Population structure.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
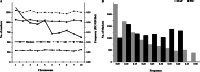
References
-
- Borlaug NE. Contributions of conventional plant breeding to food production. Science. 1983;219:689–693. - PubMed
-
- Cooper HD, Spillane C, Hodgkin T. Broadening the genetic base of crop production. Wallingford: CABI Publishing; 2001.
-
- Blaustein RJ. The green revolution arrives in Africa. Bioscience. 2008;58(1):8–14.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

