Identification of lipidomic profiles associated with drug-resistant prostate cancer cells
- PMID: 33596934
- PMCID: PMC7890620
- DOI: 10.1186/s12944-021-01437-5
Identification of lipidomic profiles associated with drug-resistant prostate cancer cells
Abstract
Background: The association of circulating lipids with clinical outcomes of drug-resistant castration-resistant prostate cancer (DR-CRPC) is not fully understood. While it is known that increases in select lipids correlate to decreased survival, neither the mechanisms mediating these alterations nor the correlation of resistance to drug treatments is well characterized.
Methods: This gap-in-knowledge was addressed using in vitro models of non-cancerous, hormone-sensitive, CRPC and drug-resistant cell lines combined with quantitative LC-ESI-Orbitrap-MS (LC-ESI-MS/MS) lipidomic analysis and subsequent analysis such as Metaboanalyst and Lipid Pathway Enrichment Analysis (LIPEA).
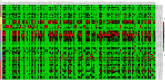
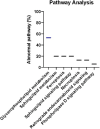
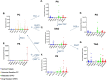
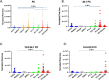
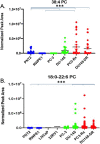
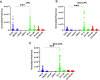
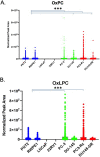
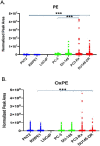
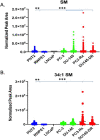
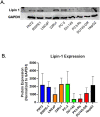
Results: Several lipid regulatory pathways were identified that are associated with Docetaxel resistance in prostate cancer (PCa). These included those controlling glycerophospholipid metabolism, sphingolipid signaling and ferroptosis. In total, 7460 features were identified as being dysregulated between the cell lines studied, and 21 lipid species were significantly altered in drug-resistant cell lines as compared to nonresistant cell lines. Docetaxel resistance cells (PC3-Rx and DU145-DR) had higher levels of phosphatidylcholine (PC), oxidized lipid species, phosphatidylethanolamine (PE), and sphingomyelin (SM) as compared to parent control cells (PC-3 and DU-145). Alterations were also identified in the levels of phosphatidic acid (PA) and diacylglyceride (DAG), whose levels are regulated by Lipin (LPIN), a phosphatidic acid phosphatase that converts PA to DAG. Data derived from cBioPortal demonstrated a population of PCa patients expressing mutations aligning with amplification of LPIN1, LPIN2 and LPIN3 genes. Lipin amplification in these genes correlated to decreased survival in these patients. Lipin-1 mRNA expression also showed a similar trend in PCa patient data. Lipin-1, but not Lipin-2 or - 3, was detected in several prostate cancer cells, and was increased in 22RV1 and PC-3 cell lines. The increased expression of Lipin-1 in these cells correlated with the level of PA.
Conclusion: These data identify lipids whose levels may correlate to Docetaxel sensitivity and progression of PCa. The data also suggest a correlation between the expression of Lipin-1 in cells and patients with regards to prostate cancer cell aggressiveness and patient survivability. Ultimately, these data may be useful for identifying markers of lethal and/or metastatic prostate cancer.
Keywords: Drug resistance; Lipid metabolism; Lipid species; Lipidomics; Lipids; Mass spectrometry; Metastasis; Prostate; Prostate Cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Identification of blood lipid markers of docetaxel treatment in prostate cancer patients.Sci Rep. 2024 Sep 27;14(1):22069. doi: 10.1038/s41598-024-73074-8. Sci Rep. 2024. PMID: 39333185 Free PMC article.
-
Lipin-1 regulates cancer cell phenotype and is a potential target to potentiate rapamycin treatment.Oncotarget. 2015 May 10;6(13):11264-80. doi: 10.18632/oncotarget.3595. Oncotarget. 2015. PMID: 25834103 Free PMC article.
-
Overexpression of claspin promotes docetaxel resistance and is associated with prostate-specific antigen recurrence in prostate cancer.Cancer Med. 2021 Aug;10(16):5574-5588. doi: 10.1002/cam4.4113. Epub 2021 Jul 9. Cancer Med. 2021. PMID: 34240817 Free PMC article.
-
Mechanisms of docetaxel resistance in prostate cancer: The key role played by miRNAs.Biochim Biophys Acta Rev Cancer. 2021 Jan;1875(1):188481. doi: 10.1016/j.bbcan.2020.188481. Epub 2020 Nov 17. Biochim Biophys Acta Rev Cancer. 2021. PMID: 33217485 Review.
-
[Research and development of Lipin family.].Yi Chuan. 2010 Oct;32(10):981-93. doi: 10.3724/sp.j.1005.2010.00981. Yi Chuan. 2010. PMID: 20943485 Review. Chinese.
Cited by
-
Recent progress of ferroptosis in cancers and drug discovery.Asian J Pharm Sci. 2024 Aug;19(4):100939. doi: 10.1016/j.ajps.2024.100939. Epub 2024 Jun 26. Asian J Pharm Sci. 2024. PMID: 39246507 Free PMC article. Review.
-
Untargeted lipidomic analysis and network pharmacology for parthenolide treated papillary thyroid carcinoma cells.BMC Complement Med Ther. 2023 Apr 24;23(1):130. doi: 10.1186/s12906-023-03944-7. BMC Complement Med Ther. 2023. PMID: 37095470 Free PMC article.
-
Dihydroceramide desaturase 1 (DES1) promotes anchorage-independent survival downstream of HER2-driven glucose uptake and metabolism.FASEB J. 2022 Oct;36(10):e22558. doi: 10.1096/fj.202200748R. FASEB J. 2022. PMID: 36165222 Free PMC article.
-
FACS-assisted single-cell lipidome analysis of phosphatidylcholines and sphingomyelins in cells of different lineages.J Lipid Res. 2023 Mar;64(3):100341. doi: 10.1016/j.jlr.2023.100341. Epub 2023 Feb 4. J Lipid Res. 2023. PMID: 36740022 Free PMC article.
-
Lipin-1, a Versatile Regulator of Lipid Homeostasis, Is a Potential Target for Fighting Cancer.Int J Mol Sci. 2021 Apr 23;22(9):4419. doi: 10.3390/ijms22094419. Int J Mol Sci. 2021. PMID: 33922580 Free PMC article. Review.
References
-
- George DJ, Kantoff PW, Lin DW. New and emerging treatments for advanced prostate cancer. Clin Adv Hematol Oncol. 2011;9(6 Suppl 12):1–15. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

