Packaging signal of influenza A virus
- PMID: 33596956
- PMCID: PMC7890907
- DOI: 10.1186/s12985-021-01504-4
Packaging signal of influenza A virus
Abstract
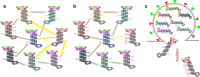
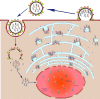
Influenza A virus (IAV) contains a genome with eight single-stranded, negative-sense RNA segments that encode 17 proteins. During its assembly, all eight separate viral RNA (vRNA) segments are incorporated into virions in a selective manner. Evidence suggested that the highly selective genome packaging mechanism relies on RNA-RNA or protein-RNA interactions. The specific structures of each vRNA that contribute to mediating the packaging of the vRNA into virions have been described and identified as packaging signals. Abundant research indicated that sequences required for genome incorporation are not series and are varied among virus genotypes. The packaging signals play important roles in determining the virus replication, genome incorporation and genetic reassortment of influenza A virus. In this review, we discuss recent studies on influenza A virus packaging signals to provide an overview of their characteristics and functions.
Keywords: Incorporation; Influenza A virus; Packaging signals; RNA-RNA interaction; Reassortment.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Contribution of RNA-RNA Interactions Mediated by the Genome Packaging Signals for the Selective Genome Packaging of Influenza A Virus.J Virol. 2022 Mar 23;96(6):e0164121. doi: 10.1128/JVI.01641-21. Epub 2022 Jan 19. J Virol. 2022. PMID: 35044211 Free PMC article.
-
RNA Sequence Features Are at the Core of Influenza A Virus Genome Packaging.J Mol Biol. 2019 Oct 4;431(21):4217-4228. doi: 10.1016/j.jmb.2019.03.018. Epub 2019 Mar 23. J Mol Biol. 2019. PMID: 30914291 Free PMC article. Review.
-
Selective Genome Packaging Mechanisms of Influenza A Viruses.Cold Spring Harb Perspect Med. 2021 Jul 1;11(7):a038497. doi: 10.1101/cshperspect.a038497. Cold Spring Harb Perspect Med. 2021. PMID: 32839251 Free PMC article. Review.
-
The genome-packaging signal of the influenza A virus genome comprises a genome incorporation signal and a genome-bundling signal.J Virol. 2013 Nov;87(21):11316-22. doi: 10.1128/JVI.01301-13. Epub 2013 Aug 7. J Virol. 2013. PMID: 23926345 Free PMC article.
-
Influenza A and B virus intertypic reassortment through compatible viral packaging signals.J Virol. 2014 Sep;88(18):10778-91. doi: 10.1128/JVI.01440-14. Epub 2014 Jul 9. J Virol. 2014. PMID: 25008914 Free PMC article.
Cited by
-
Recent Insights into the Molecular Mechanisms of the Toll-like Receptor Response to Influenza Virus Infection.Int J Mol Sci. 2024 May 29;25(11):5909. doi: 10.3390/ijms25115909. Int J Mol Sci. 2024. PMID: 38892096 Free PMC article. Review.
-
Global research trends in the relationship between influenza and CD4+ T/CD8+ T cells: A bibliometric analysis.Hum Vaccin Immunother. 2024 Dec 31;20(1):2435644. doi: 10.1080/21645515.2024.2435644. Epub 2024 Dec 16. Hum Vaccin Immunother. 2024. PMID: 39680034 Free PMC article.
-
Reciprocal enhancement of SARS-CoV-2 and influenza virus replication in human pluripotent stem cell-derived lung organoids1.Emerg Microbes Infect. 2023 Dec;12(1):2211685. doi: 10.1080/22221751.2023.2211685. Emerg Microbes Infect. 2023. PMID: 37161660 Free PMC article.
-
Nuclear AGO2 supports influenza A virus replication through type-I interferon regulation.Nucleic Acids Res. 2025 Apr 10;53(7):gkaf268. doi: 10.1093/nar/gkaf268. Nucleic Acids Res. 2025. PMID: 40219968 Free PMC article.
-
Revisiting influenza A virus life cycle from a perspective of genome balance.Virol Sin. 2023 Feb;38(1):1-8. doi: 10.1016/j.virs.2022.10.005. Epub 2022 Oct 27. Virol Sin. 2023. PMID: 36309307 Free PMC article. Review.
References
-
- Saumyadip Sarkar BPT. Cover Story: Influenza D - New Virus Classified. We The Microbiologist. 2016;3:24–26.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

