Intranasal fusion inhibitory lipopeptide prevents direct-contact SARS-CoV-2 transmission in ferrets
- PMID: 33597220
- PMCID: PMC8011693
- DOI: 10.1126/science.abf4896
Intranasal fusion inhibitory lipopeptide prevents direct-contact SARS-CoV-2 transmission in ferrets
Abstract
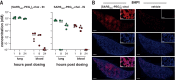
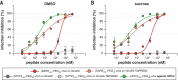
Containment of the COVID-19 pandemic requires reducing viral transmission. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is initiated by membrane fusion between the viral and host cell membranes, which is mediated by the viral spike protein. We have designed lipopeptide fusion inhibitors that block this critical first step of infection and, on the basis of in vitro efficacy and in vivo biodistribution, selected a dimeric form for evaluation in an animal model. Daily intranasal administration to ferrets completely prevented SARS-CoV-2 direct-contact transmission during 24-hour cohousing with infected animals, under stringent conditions that resulted in infection of 100% of untreated animals. These lipopeptides are highly stable and thus may readily translate into safe and effective intranasal prophylaxis to reduce transmission of SARS-CoV-2.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures


Update of
-
Intranasal fusion inhibitory lipopeptide prevents direct contact SARS-CoV-2 transmission in ferrets.bioRxiv [Preprint]. 2020 Nov 5:2020.11.04.361154. doi: 10.1101/2020.11.04.361154. bioRxiv. 2020. Update in: Science. 2021 Mar 26;371(6536):1379-1382. doi: 10.1126/science.abf4896. PMID: 33173865 Free PMC article. Updated. Preprint.
References
-
- Hoffmann M., Kleine-Weber H., Schroeder S., Krger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N.-H., Nitsche A., Mller M. A., Drosten C., Phlmann S., SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8. (2020). 10.1016/j.cell.2020.02.052 - DOI - PMC - PubMed
-
- Outlaw V. K., Bovier F. T., Mears M. C., Cajimat M. N., Zhu Y., Lin M. J., Addetia A., Lieberman N. A. P., Peddu V., Xie X., Shi P.-Y., Greninger A. L., Gellman S. H., Bente D. A., Moscona A., Porotto M., Inhibition of coronavirus entry in vitro and ex vivo by a lipid-conjugated peptide derived from the SARS-CoV-2 spike glycoprotein HRC domain. mBio 11, e01935-20 (2020). 10.1128/mBio.01935-20 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

