Microfluidic tumor-on-a-chip model to evaluate the role of tumor environmental stress on NK cell exhaustion
- PMID: 33597234
- PMCID: PMC7888951
- DOI: 10.1126/sciadv.abc2331
Microfluidic tumor-on-a-chip model to evaluate the role of tumor environmental stress on NK cell exhaustion
Abstract
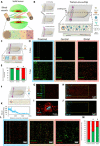
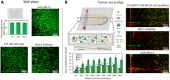
Solid tumors generate a suppressive environment that imposes an overwhelming burden on the immune system. Nutrient depletion, waste product accumulation, hypoxia, and pH acidification severely compromise the capacity of effector immune cells such as T and natural killer (NK) cells to destroy cancer cells. However, the specific molecular mechanisms driving immune suppression, as well as the capacity of immune cells to adapt to the suppressive environment, are not completely understood. Thus, here, we used an in vitro microfluidic tumor-on-a-chip platform to evaluate how NK cells respond to the tumor-induced suppressive environment. The results demonstrated that the suppressive environment created by the tumor gradually eroded NK cell cytotoxic capacity, leading to compromised NK cell surveillance and tumor tolerance. Further, NK cell exhaustion persisted for an extended period of time after removing NK cells from the microfluidic platform. Last, the addition of checkpoint inhibitors and immunomodulatory agents alleviated NK cell exhaustion.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures





References
-
- Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30 (2020). - PubMed
-
- Schultz L., Mackall C., Driving CAR T cell translation forward. Sci. Transl. Med. 11, eaaw2127 (2019). - PubMed
-
- Mardiana S., Solomon B. J., Darcy P. K., Beavis P. A., Supercharging adoptive T cell therapy to overcome solid tumor–induced immunosuppression. Sci. Transl. Med. 11, eaaw2293 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

