Analysis of recurrently protected genomic regions in cell-free DNA found in urine
- PMID: 33597261
- PMCID: PMC9258975
- DOI: 10.1126/scitranslmed.aaz3088
Analysis of recurrently protected genomic regions in cell-free DNA found in urine
Abstract
Cell-free DNA (cfDNA) in urine is a promising analyte for noninvasive diagnostics. However, urine cfDNA is highly fragmented. Whether characteristics of these fragments reflect underlying genomic architecture is unknown. Here, we characterized fragmentation patterns in urine cfDNA using whole-genome sequencing. Size distribution of urine cfDNA fragments showed multiple strong peaks between 40 and 120 base pairs (bp) with a modal size of 81- and sharp 10-bp periodicity, suggesting transient protection from complete degradation. These properties were robust to preanalytical perturbations, such as at-home collection and delay in processing. Genome-wide sequencing coverage of urine cfDNA fragments revealed recurrently protected regions (RPRs) conserved across individuals, with partial overlap with nucleosome positioning maps inferred from plasma cfDNA. The ends of cfDNA fragments clustered upstream and downstream of RPRs, and nucleotide frequencies of fragment ends indicated enzymatic digestion of urine cfDNA. Compared to plasma, fragmentation patterns in urine cfDNA showed greater correlation with gene expression and chromatin accessibility in epithelial cells of the urinary tract. We determined that tumor-derived urine cfDNA exhibits a higher frequency of aberrant fragments that end within RPRs. By comparing the fraction of aberrant fragments and nucleotide frequencies of fragment ends, we identified urine samples from cancer patients with an area under the curve of 0.89. Our results revealed nonrandom genomic positioning of urine cfDNA fragments and suggested that analysis of fragmentation patterns across recurrently protected genomic loci may serve as a cancer diagnostic.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures
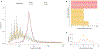


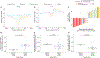




References
-
- Wong FC, Lo YM, Prenatal Diagnosis Innovation: Genome Sequencing of Maternal Plasma. Annu Rev Med 67, 419–432 (2016). - PubMed
-
- Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, Pacey S, Baird R, Rosenfeld N, Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 17, 223–238 (2017). - PubMed
-
- Murtaza M, Caldas C, Nucleosome mapping in plasma DNA predicts cancer gene expression. Nat Genet 48, 1105–1106 (2016). - PubMed
-
- Cristiano S, Leal A, Phallen J, Fiksel J, Adleff V, Bruhm DC, Jensen SO, Medina JE, Hruban C, White JR, Palsgrove DN, Niknafs N, Anagnostou V, Forde P, Naidoo J, Marrone K, Brahmer J, Woodward BD, Husain H, van Rooijen KL, Orntoft MW, Madsen AH, van de Velde CJH, Verheij M, Cats A, Punt CJA, Vink GR, van Grieken NCT, Koopman M, Fijneman RJA, Johansen JS, Nielsen HJ, Meijer GA, Andersen CL, Scharpf RB, Velculescu VE, Genome-wide cell-free DNA fragmentation in patients with cancer. Nature, (2019). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

