A tetravalent live attenuated dengue virus vaccine stimulates balanced immunity to multiple serotypes in humans
- PMID: 33597521
- PMCID: PMC7889627
- DOI: 10.1038/s41467-021-21384-0
A tetravalent live attenuated dengue virus vaccine stimulates balanced immunity to multiple serotypes in humans
Abstract
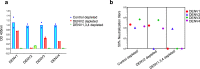
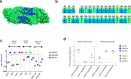
The four-dengue virus (DENV) serotypes infect several hundred million people annually. For the greatest safety and efficacy, tetravalent DENV vaccines are designed to stimulate balanced protective immunity to all four serotypes. However, this has been difficult to achieve. Clinical trials with a leading vaccine demonstrated that unbalanced replication and immunodominance of one vaccine component over others can lead to low efficacy and vaccine enhanced severe disease. The Laboratory of Infectious Diseases at the National Institutes of Health has developed a live attenuated tetravalent DENV vaccine (TV003), which is currently being tested in phase 3 clinical trials. Here we report, our study to determine if TV003 stimulate balanced and serotype-specific (TS) neutralizing antibody (nAb) responses to each serotype. Serum samples from twenty-one dengue-naive individuals participated under study protocol CIR287 (ClinicalTrials.gov NCT02021968) are analyzed 6 months after vaccination. Most subjects (76%) develop TS nAbs to 3 or 4 DENV serotypes, indicating immunity is induced by each vaccine component. Vaccine-induced TS nAbs map to epitopes known to be targets of nAbs in people infected with wild type DENVs. Following challenge with a partially attenuated strain of DENV2, all 21 subjects are protected from the efficacy endpoints. However, some vaccinated individuals develop post challenge nAb boost, while others mount post-challenge antibody responses that are consistent with sterilizing immunity. TV003 vaccine induced DENV2 TS nAbs are associated with sterilizing immunity. Our results indicate that nAbs to TS epitopes on each serotype may be a better correlate than total levels of nAbs currently used for guiding DENV vaccine development.
Conflict of interest statement
A.M.D. has consulted on Dengue vaccine for Takeda vaccines, Sanofi Pasteur, GSK, and Merck Pharmaceuticals and also an inventor in patents related to Dengue vaccines. R.S.B. has consulted on Dengue vaccines for Takeda Vaccines and Sanofi Pasteur and is also an inventor in patents related to Dengue vaccines. All other authors report no potential conflicts of interest.
Figures





References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

