Identification of novel cell glycolysis related gene signature predicting survival in patients with breast cancer
- PMID: 33597614
- PMCID: PMC7889867
- DOI: 10.1038/s41598-021-83628-9
Identification of novel cell glycolysis related gene signature predicting survival in patients with breast cancer
Abstract
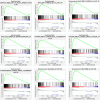
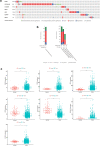
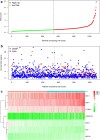
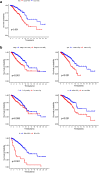
One of the most frequently identified tumors and a contributing cause of death in women is breast cancer (BC). Many biomarkers associated with survival and prognosis were identified in previous studies through database mining. Nevertheless, the predictive capabilities of single-gene biomarkers are not accurate enough. Genetic signatures can be an enhanced prediction method. This research analyzed data from The Cancer Genome Atlas (TCGA) for the detection of a new genetic signature to predict BC prognosis. Profiling of mRNA expression was carried out in samples of patients with TCGA BC (n = 1222). Gene set enrichment research has been undertaken to classify gene sets that vary greatly between BC tissues and normal tissues. Cox models for additive hazards regression were used to classify genes that were strongly linked to overall survival. A subsequent Cox regression multivariate analysis was used to construct a predictive risk parameter model. Kaplan-Meier survival predictions and log-rank validation have been used to verify the value of risk prediction parameters. Seven genes (PGK1, CACNA1H, IL13RA1, SDC1, AK3, NUP43, SDC3) correlated with glycolysis were shown to be strongly linked to overall survival. Depending on the 7-gene-signature, 1222 BC patients were classified into subgroups of high/low-risk. Certain variables have not impaired the prognostic potential of the seven-gene signature. A seven-gene signature correlated with cellular glycolysis was developed to predict the survival of BC patients. The results include insight into cellular glycolysis mechanisms and the detection of patients with poor BC prognosis.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Identification of a novel glycolysis-related signature to predict the prognosis of patients with breast cancer.World J Surg Oncol. 2021 Oct 2;19(1):294. doi: 10.1186/s12957-021-02409-w. World J Surg Oncol. 2021. PMID: 34600547 Free PMC article.
-
A novel glycolysis-related gene signature for predicting prognosis and immunotherapy efficacy in breast cancer.Front Immunol. 2025 Feb 19;16:1512859. doi: 10.3389/fimmu.2025.1512859. eCollection 2025. Front Immunol. 2025. PMID: 40046063 Free PMC article.
-
Identification and prognostic value of a glycolysis-related gene signature in patients with bladder cancer.Medicine (Baltimore). 2021 Jan 22;100(3):e23836. doi: 10.1097/MD.0000000000023836. Medicine (Baltimore). 2021. PMID: 33545950 Free PMC article.
-
Molecular and clinical profiles of syndecan-1 in solid and hematological cancer for prognosis and precision medicine.Oncotarget. 2015 Oct 6;6(30):28693-715. doi: 10.18632/oncotarget.4981. Oncotarget. 2015. PMID: 26293675 Free PMC article. Review.
-
New Advances in Molecular Breast Cancer Pathology.Semin Cancer Biol. 2021 Jul;72:102-113. doi: 10.1016/j.semcancer.2020.03.014. Epub 2020 Apr 5. Semin Cancer Biol. 2021. PMID: 32259641 Review.
Cited by
-
Prognostic value of amino acid metabolism-related gene expression in invasive breast carcinoma.J Cancer Res Clin Oncol. 2023 Oct;149(13):11117-11133. doi: 10.1007/s00432-023-04985-8. Epub 2023 Jun 21. J Cancer Res Clin Oncol. 2023. PMID: 37340191 Free PMC article.
-
Increased tumor glycolysis is associated with decreased immune infiltration across human solid tumors.Front Immunol. 2022 Nov 24;13:880959. doi: 10.3389/fimmu.2022.880959. eCollection 2022. Front Immunol. 2022. PMID: 36505421 Free PMC article.
-
Effects of Glycolysis-Related Genes on Prognosis and the Tumor Microenvironment of Hepatocellular Carcinoma.Front Pharmacol. 2022 Jul 18;13:895608. doi: 10.3389/fphar.2022.895608. eCollection 2022. Front Pharmacol. 2022. PMID: 35924040 Free PMC article.
-
Crocodylus porosus Sera a Potential Source to Identify Novel Epigenetic Targets: In Silico Analysis.Vet Sci. 2022 Apr 25;9(5):210. doi: 10.3390/vetsci9050210. Vet Sci. 2022. PMID: 35622738 Free PMC article.
-
Facts and Perspectives: Implications of tumor glycolysis on immunotherapy response in triple negative breast cancer.Front Oncol. 2023 Jan 10;12:1061789. doi: 10.3389/fonc.2022.1061789. eCollection 2022. Front Oncol. 2023. PMID: 36703796 Free PMC article. Review.
References
-
- Schwartz AM, Henson DE, Chen D, Rajamarthandan S. Histologic grade remains a prognostic factor for breast cancer regardless of the number of positive lymph nodes and tumor size: a study of 161 708 cases of breast cancer from the SEER Program. Arch. Pathol. Lab. Med. 2014;138:1048. doi: 10.5858/arpa.2013-0435-OA. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

