SIRT1 and SIRT2 Activity Control in Neurodegenerative Diseases
- PMID: 33597872
- PMCID: PMC7883599
- DOI: 10.3389/fphar.2020.585821
SIRT1 and SIRT2 Activity Control in Neurodegenerative Diseases
Abstract
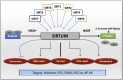
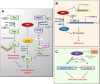
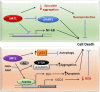
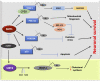
Sirtuins are NAD+ dependent histone deacetylases (HDAC) that play a pivotal role in neuroprotection and cellular senescence. SIRT1-7 are different homologs from sirtuins. They play a prominent role in many aspects of physiology and regulate crucial proteins. Modulation of sirtuins can thus be utilized as a therapeutic target for metabolic disorders. Neurological diseases have distinct clinical manifestations but are mainly age-associated and due to loss of protein homeostasis. Sirtuins mediate several life extension pathways and brain functions that may allow therapeutic intervention for age-related diseases. There is compelling evidence to support the fact that SIRT1 and SIRT2 are shuttled between the nucleus and cytoplasm and perform context-dependent functions in neurodegenerative diseases including Alzheimer's disease (AD), Parkinson's disease (PD), and Huntington's disease (HD). In this review, we highlight the regulation of SIRT1 and SIRT2 in various neurological diseases. This study explores the various modulators that regulate the activity of SIRT1 and SIRT2, which may further assist in the treatment of neurodegenerative disease. Moreover, we analyze the structure and function of various small molecules that have potential significance in modulating sirtuins, as well as the technologies that advance the targeted therapy of neurodegenerative disease.
Keywords: SIRT1; SIRT2; modulators; neurodegenerative diseases; neuroprotective mechanism; resveratrol; selective pockets, sir reals.
Copyright © 2021 Manjula, Anuja and Alcain.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
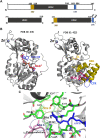
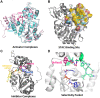

Similar articles
-
SIRT1 and SIRT2: emerging targets in neurodegeneration.EMBO Mol Med. 2013 Mar;5(3):344-52. doi: 10.1002/emmm.201302451. Epub 2013 Feb 18. EMBO Mol Med. 2013. PMID: 23417962 Free PMC article. Review.
-
The Role of Sirtuin 1 (SIRT1) in Neurodegeneration.Cureus. 2023 Jun 15;15(6):e40463. doi: 10.7759/cureus.40463. eCollection 2023 Jun. Cureus. 2023. PMID: 37456463 Free PMC article. Review.
-
Sirtuins and Their Roles in Brain Aging and Neurodegenerative Disorders.Neurochem Res. 2017 Mar;42(3):876-890. doi: 10.1007/s11064-016-2110-y. Epub 2016 Nov 24. Neurochem Res. 2017. PMID: 27882448 Free PMC article. Review.
-
Sirtuins as possible targets in neurodegenerative diseases.Curr Drug Targets. 2013 Jun 1;14(6):644-7. doi: 10.2174/1389450111314060004. Curr Drug Targets. 2013. PMID: 23410123 Review.
-
Review of the anti-inflammatory effect of SIRT1 and SIRT2 modulators on neurodegenerative diseases.Eur J Pharmacol. 2020 Jan 15;867:172847. doi: 10.1016/j.ejphar.2019.172847. Epub 2019 Dec 5. Eur J Pharmacol. 2020. PMID: 31812544 Review.
Cited by
-
Sirt1: An Increasingly Interesting Molecule with a Potential Role in Bone Metabolism and Osteoporosis.Biomolecules. 2024 Aug 8;14(8):970. doi: 10.3390/biom14080970. Biomolecules. 2024. PMID: 39199358 Free PMC article. Review.
-
Editorial: Allosteric functions and inhibitions: structural insights.Front Mol Biosci. 2024 Jan 15;11:1363100. doi: 10.3389/fmolb.2024.1363100. eCollection 2024. Front Mol Biosci. 2024. PMID: 38293599 Free PMC article. No abstract available.
-
Exploring the Multi-Faceted Role of Sirtuins in Glioblastoma Pathogenesis and Targeting Options.Int J Mol Sci. 2022 Oct 25;23(21):12889. doi: 10.3390/ijms232112889. Int J Mol Sci. 2022. PMID: 36361680 Free PMC article. Review.
-
Resveratrol and neuroprotection: an insight into prospective therapeutic approaches against Alzheimer's disease from bench to bedside.Mol Neurobiol. 2022 Jul;59(7):4384-4404. doi: 10.1007/s12035-022-02859-7. Epub 2022 May 12. Mol Neurobiol. 2022. PMID: 35545730 Review.
-
Efficient Assay and Marker Significance of NAD+ in Human Blood.Front Med (Lausanne). 2022 May 19;9:886485. doi: 10.3389/fmed.2022.886485. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35665345 Free PMC article.
References
-
- Albani D., Polito L., Batelli S., De Mauro S., Fracasso C., Martelli G., et al. (2009). The SIRT1 activator resveratrol protects SK-N-BE cells from oxidative stress and against toxicity caused by alpha-synuclein or amyloid-beta (1-42) peptide. J. Neurochem. 110, 1445–1456. 10.1111/j.1471-4159.2009.06228.x - DOI - PubMed
-
- Alcaín F. J., Domínguez J., Durán-Prado M., Vaamonde J. (2020). “Coenzyme Q and age‐related neurodegenerative disorders: Parkinson and Alzheimer diseases BT,” in Coenzyme Q in aging. Editor López Lluch G. (Cham: Springer International Publishing; ), 241–268.
-
- Arunsundar M., Shanmugarajan T. S., Ravichandran V. (2014). 3,4-dihydroxyphenylethanol attenuates spatio-cognitive deficits in an alzheimer’s disease mouse model: modulation of the molecular signals in neuronal survival-apoptotic programs. Neurotox. Res. 27, 143–155. 10.1007/s12640-014-9492-x - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

