Developing Innolysins Against Campylobacter jejuni Using a Novel Prophage Receptor-Binding Protein
- PMID: 33597938
- PMCID: PMC7882524
- DOI: 10.3389/fmicb.2021.619028
Developing Innolysins Against Campylobacter jejuni Using a Novel Prophage Receptor-Binding Protein
Abstract
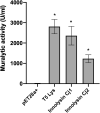
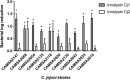
Campylobacter contaminated poultry remains the major cause of foodborne gastroenteritis worldwide, calling for novel antibacterials. We previously developed the concept of Innolysin composed of an endolysin fused to a phage receptor binding protein (RBP) and provided the proof-of-concept that Innolysins exert bactericidal activity against Escherichia coli. Here, we have expanded the Innolysin concept to target Campylobacter jejuni. As no C. jejuni phage RBP had been identified so far, we first showed that the H-fiber originating from a CJIE1-like prophage of C. jejuni CAMSA2147 functions as a novel RBP. By fusing this H-fiber to phage T5 endolysin, we constructed Innolysins targeting C. jejuni (Innolysins Cj). Innolysin Cj1 exerts antibacterial activity against diverse C. jejuni strains after in vitro exposure for 45 min at 20°C, reaching up to 1.30 ± 0.21 log reduction in CAMSA2147 cell counts. Screening of a library of Innolysins Cj composed of distinct endolysins for growth inhibition, allowed us to select Innolysin Cj5 as an additional promising antibacterial candidate. Application of either Innolysin Cj1 or Innolysin Cj5 on chicken skin refrigerated to 5°C and contaminated with C. jejuni CAMSA2147 led to 1.63 ± 0.46 and 1.18 ± 0.10 log reduction of cells, respectively, confirming that Innolysins Cj can kill C. jejuni in situ. The receptor of Innolysins Cj remains to be identified, however, the RBP component (H-fiber) recognizes a novel receptor compared to lytic phages binding to capsular polysaccharide or flagella. Identification of other unexplored Campylobacter phage RBPs may further increase the repertoire of new Innolysins Cj targeting distinct receptors and working as antibacterials against Campylobacter.
Keywords: Campylobacter; Innolysin; antibacterials; endolysin; food safety; prophage binding.
Copyright © 2021 Zampara, Sørensen, Gencay, Grimon, Kristiansen, Jørgensen, Kristensen, Briers, Elsser-Gravesen and Brøndsted.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Exploiting phage receptor binding proteins to enable endolysins to kill Gram-negative bacteria.Sci Rep. 2020 Jul 21;10(1):12087. doi: 10.1038/s41598-020-68983-3. Sci Rep. 2020. PMID: 32694655 Free PMC article.
-
Campycins are novel broad-spectrum antibacterials killing Campylobacter jejuni.Appl Microbiol Biotechnol. 2024 Oct 9;108(1):484. doi: 10.1007/s00253-024-13317-w. Appl Microbiol Biotechnol. 2024. PMID: 39382702 Free PMC article.
-
A hybrid receptor binding protein enables phage F341 infection of Campylobacter by binding to flagella and lipooligosaccharides.Front Microbiol. 2024 Feb 1;15:1358909. doi: 10.3389/fmicb.2024.1358909. eCollection 2024. Front Microbiol. 2024. PMID: 38380094 Free PMC article.
-
Characterization and Application of Lytic Bacteriophages against Campylobacter jejuni Isolated from Poultry in Japan.Biocontrol Sci. 2017;22(4):213-221. doi: 10.4265/bio.22.213. Biocontrol Sci. 2017. PMID: 29279578
-
Application of Campylobacter jejuni Phages: Challenges and Perspectives.Animals (Basel). 2020 Feb 11;10(2):279. doi: 10.3390/ani10020279. Animals (Basel). 2020. PMID: 32054081 Free PMC article. Review.
Cited by
-
A comprehensive review of the applications of bacteriophage-derived endolysins for foodborne bacterial pathogens and food safety: recent advances, challenges, and future perspective.Front Microbiol. 2023 Oct 6;14:1259210. doi: 10.3389/fmicb.2023.1259210. eCollection 2023. Front Microbiol. 2023. PMID: 37869651 Free PMC article. Review.
-
Genomic analysis and characterization of lytic bacteriophages that target antimicrobial resistant Escherichia coli in Addis Ababa, Ethiopia.Heliyon. 2024 Nov 12;10(22):e40342. doi: 10.1016/j.heliyon.2024.e40342. eCollection 2024 Nov 30. Heliyon. 2024. PMID: 39619596 Free PMC article.
-
Engineering an antimicrobial chimeric endolysin that targets the phytopathogen Pseudomonas syringae pv. actinidiae.J Biol Chem. 2025 Jun;301(6):110224. doi: 10.1016/j.jbc.2025.110224. Epub 2025 May 9. J Biol Chem. 2025. PMID: 40349779 Free PMC article.
-
Isolation and Characterization of a Novel Virulent Phage ASG01 of Aeromonas salmonicida and Its Cell Wall Hydrolase Activity.Microorganisms. 2024 Mar 21;12(3):629. doi: 10.3390/microorganisms12030629. Microorganisms. 2024. PMID: 38543679 Free PMC article.
-
ECOPHAGE: Combating Antimicrobial Resistance Using Bacteriophages for Eco-Sustainable Agriculture and Food Systems.Viruses. 2023 Nov 8;15(11):2224. doi: 10.3390/v15112224. Viruses. 2023. PMID: 38005900 Free PMC article.
References
-
- Anjum A., Brathwaite K. J., Aidley J., Connerton P. L., Cummings N. J., Parkhill J., et al. (2016). Phase variation of a Type IIG restriction-modification enzyme alters site-specific methylation patterns and gene expression in Campylobacter jejuni strain NCTC11168. Nucleic Acids Res. 44 4581–4594. 10.1093/nar/gkw019 - DOI - PMC - PubMed
-
- Biesta-Peters E. G., Jongenburger I., de Boer E., Jacobs-Reitsma W. F. (2019). Validation by interlaboratory trials of EN ISO 10272 - Microbiology of the food chain – Horizontal method for detection and enumeration of Campylobacter spp. – Part 1: detection method. Int. J. Food Microbiol. 288 39–46. 10.1016/j.ijfoodmicro.2018.05.007 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

