Identification of Serum Circulating MicroRNAs as Novel Diagnostic Biomarkers of Gastric Cancer
- PMID: 33597967
- PMCID: PMC7882724
- DOI: 10.3389/fgene.2020.591515
Identification of Serum Circulating MicroRNAs as Novel Diagnostic Biomarkers of Gastric Cancer
Abstract
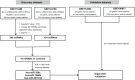
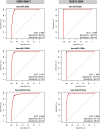
Gastric cancer (GC) is one of the leading causes of cancer-associated deaths worldwide. Due to the lack of typical symptoms and effective biomarkers for non-invasive screening, most patients develop advanced-stage GC by the time of diagnosis. Circulating microRNA (miRNA)-based panels have been reported as a promising tool for the screening of certain types of cancers. In this study, we performed differential expression analysis of miRNA profiles of plasma samples obtained from gastric cancer and non-cancer patients using two independent Gene Expression Omnibus (GEO) datasets: GSE113486 and GSE124158. We identified three miRNAs, hsa-miR-320a, hsa-miR-1260b, and hsa-miR-6515-5p, to distinguish gastric cancer cases from non-cancer controls. The three miRNAs showed an area under the curve (AUC) over 0.95 with high specificity (>93.0%) and sensitivity (>85.0%) in both the discovery datasets. In addition, we further validated these three miRNAs in two external datasets: GSE106817 [sensitivity: hsa-miR-320a (99.1%), hsa-miR-1260b (97.4%), and hsa-miR-6515-5p (92.2%); specificity: hsa-miR-320a (88.8%), hsa-miR-1260b (89.6%), and hsa-miR-6515-5p (88.7%); and AUC: hsa-miR-320a (96.3%), hsa-miR-1260b (97.4%), and hsa-miR-6515-5p (94.6%)] and GSE112264 [sensitivity: hsa-miR-320a (100.0%), hsa-miR-1260b (98.0%), and hsa-miR-6515.5p (98.0%); specificity: hsa-miR-320a (100.0%), hsa-miR-1260b (100.0%), and hsa-miR-6515.5p (92.7%); and AUC: hsa-miR-320a (1.000), hsa-miR-1260b (1.000), and hsa-miR-6515-5p (0.988)]. On the basis of these findings, the three miRNAs can be used as potential biomarkers for gastric cancer screening, which can provide patients with a higher chance of curative resection and longer survival.
Keywords: biomarkers; differential expression analysis; gastric cancer; gene expression omnibus datasets; microRNAs.
Copyright © 2021 Yao, Ding, Bai, Zhou, Lee, Li and Teng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Identifying potential circulating miRNA biomarkers for the diagnosis and prediction of ovarian cancer using machine-learning approach: application of Boruta.Front Digit Health. 2023 Aug 9;5:1187578. doi: 10.3389/fdgth.2023.1187578. eCollection 2023. Front Digit Health. 2023. PMID: 37621964 Free PMC article.
-
Identification of non-invasive biomarkers for chronic atrophic gastritis from serum exosomal microRNAs.BMC Cancer. 2019 Feb 8;19(1):129. doi: 10.1186/s12885-019-5328-7. BMC Cancer. 2019. PMID: 30736753 Free PMC article.
-
Serum MiRNA panel as potential biomarkers for chronic hepatitis B with persistently normal alanine aminotransferase.Clin Chim Acta. 2015 Dec 7;451(Pt B):232-9. doi: 10.1016/j.cca.2015.10.002. Epub 2015 Oct 17. Clin Chim Acta. 2015. PMID: 26483130
-
Diagnostic performance of microRNAs in the detection of heart failure with reduced or preserved ejection fraction: a systematic review and meta-analysis.Eur J Heart Fail. 2022 Dec;24(12):2212-2225. doi: 10.1002/ejhf.2700. Epub 2022 Oct 27. Eur J Heart Fail. 2022. PMID: 36161443 Free PMC article.
-
Gastric Cancer MicroRNAs Meta-signature.Int J Mol Cell Med. 2019 Spring;8(2):94-102. doi: 10.22088/IJMCM.BUMS.8.2.94. Epub 2019 Aug 24. Int J Mol Cell Med. 2019. PMID: 32215261 Free PMC article. Review.
Cited by
-
MicroRNAs associated with postoperative outcomes in patients with limited stage neuroendocrine carcinoma of the esophagus.Oncol Lett. 2023 May 12;26(1):276. doi: 10.3892/ol.2023.13862. eCollection 2023 Jul. Oncol Lett. 2023. PMID: 37274462 Free PMC article.
-
Role of the TLR signaling pathway in the pathogenesis of glioblastoma multiforme with an emphasis on immunotherapy.Biochem Biophys Rep. 2025 Jul 18;43:102149. doi: 10.1016/j.bbrep.2025.102149. eCollection 2025 Sep. Biochem Biophys Rep. 2025. PMID: 40727443 Free PMC article. Review.
-
MicroRNAs miR-584-5p and miR-425-3p Are Up-Regulated in Plasma of Colorectal Cancer (CRC) Patients: Targeting with Inhibitor Peptide Nucleic Acids Is Associated with Induction of Apoptosis in Colon Cancer Cell Lines.Cancers (Basel). 2022 Dec 25;15(1):128. doi: 10.3390/cancers15010128. Cancers (Basel). 2022. PMID: 36612125 Free PMC article.
-
AGT May Serve as a Prognostic Biomarker and Correlated with Immune Infiltration in Gastric Cancer.Int J Gen Med. 2022 Feb 22;15:1865-1878. doi: 10.2147/IJGM.S351662. eCollection 2022. Int J Gen Med. 2022. PMID: 35264871 Free PMC article.
-
Novel biomarkers for early detection of gastric cancer.World J Gastroenterol. 2023 May 7;29(17):2515-2533. doi: 10.3748/wjg.v29.i17.2515. World J Gastroenterol. 2023. PMID: 37213407 Free PMC article. Review.
References
-
- Choi I. J., Lee J. H., Kim Y. I., Kim C. G., Cho S. J., Lee J. Y., et al. (2015). Long-term outcome comparison of endoscopic resection and surgery in early gastric cancer meeting the absolute indication for endoscopic resection. Gastrointest Endosc. 81 333–341. 10.1016/j.gie.2014.07.047 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

