Analysis of microbial diversity and functional differences in different types of high-temperature Daqu
- PMID: 33598183
- PMCID: PMC7866569
- DOI: 10.1002/fsn3.2068
Analysis of microbial diversity and functional differences in different types of high-temperature Daqu
Abstract
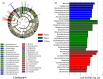
Bacterial communities that enrich in high-temperature Daqu are important for the Chinese maotai-flavor liquor brewing process. However, the bacterial communities in three different types of high-temperature Daqu (white Daqu, black Daqu, and yellow Daqu) are still undercharacterized. In this study, the bacterial diversity of three different types of high-temperature Daqu was investigated using Illumina MiSeq high-throughput sequencing. The bacterial community of high-temperature Daqu is mainly composed of thermophilic bacteria, and seven bacterial phyla along with 262 bacterial genera were identified in all 30 high-temperature Daqu samples. Firmicutes, Actinobacteria, Proteobacteria, and Acidobacteria were the dominant bacterial phyla in high-temperature Daqu samples, while Thermoactinomyces, Staphylococcus, Lentibacillus, Bacillus, Kroppenstedtia, Saccharopolyspora, Streptomyces, and Brevibacterium were the dominant bacterial genera. The bacterial community structure of three different types of high-temperature Daqu was significantly different (p < .05). In addition, the results of microbiome phenotype prediction by BugBase and bacterial functional potential prediction using PICRUSt show that bacteria from different types of high-temperature Daqu have similar functions as well as phenotypes, and bacteria in high-temperature Daqu have vigorous metabolism in the transport and decomposition of amino acids and carbohydrates. These results offer a reference for the comprehensive understanding of bacterial diversity of high-temperature Daqu.
Keywords: Chinese Maotai‐flavor liquor; Illumina MiSeq high‐throughput sequencing; bacterial diversity; functional prediction; high‐temperature Daqu.
© 2020 The Authors. Food Science & Nutrition published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Bruno, A. , Sandionigi, A. , Galimberti, A. , Siani, E. , Labra, M. , Cocuzza, C. , Ferri, E. , & Casiraghi, M. J. M. (2017). One step forwards for the routine use of high‐throughput DNA sequencing in environmental monitoring. An efficient and standardizable method to maximize the detection of environmental bacteria. MicrobiologyOpen, 6(1), e00421 10.1002/mbo3.421 - DOI - PMC - PubMed
-
- Caporaso, J. G. , Lauber, C. L. , Walters, W. A. , Berg‐Lyons, D. , Huntley, J. , Fierer, N. , Owens, S. M. , Betley, J. , Fraser, L. , Bauer, M. , Gormley, N. , Gilbert, J. A. , Smith, G. , & Knight, R. (2012). Ultra‐high‐throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME Journal, 6(8), 1621 10.1002/mbo3.421 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

