Protein Stability Perturbation Contributes to the Loss of Function in Haploinsufficient Genes
- PMID: 33598480
- PMCID: PMC7882701
- DOI: 10.3389/fmolb.2021.620793
Protein Stability Perturbation Contributes to the Loss of Function in Haploinsufficient Genes
Abstract
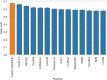
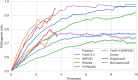
Missense variants are among the most studied genome modifications as disease biomarkers. It has been shown that the "perturbation" of the protein stability upon a missense variant (in terms of absolute ΔΔG value, i.e., |ΔΔG|) has a significant, but not predictive, correlation with the pathogenicity of that variant. However, here we show that this correlation becomes significantly amplified in haploinsufficient genes. Moreover, the enrichment of pathogenic variants increases at the increasing protein stability perturbation value. These findings suggest that protein stability perturbation might be considered as a potential cofactor in diseases associated with haploinsufficient genes reporting missense variants.
Keywords: haploinsuffciency; protein mutation; protein stability; protein stability prediction; variant effect prediction.
Copyright © 2021 Birolo, Benevenuta, Fariselli, Capriotti, Giorgio and Sanavia.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources

