Improved Quantification of Myocardium Scar in Late Gadolinium Enhancement Images: Deep Learning Based Image Fusion Approach
- PMID: 33599043
- PMCID: PMC8359184
- DOI: 10.1002/jmri.27555
Improved Quantification of Myocardium Scar in Late Gadolinium Enhancement Images: Deep Learning Based Image Fusion Approach
Abstract
Background: Quantification of myocardium scarring in late gadolinium enhanced (LGE) cardiac magnetic resonance imaging can be challenging due to low scar-to-background contrast and low image quality. To resolve ambiguous LGE regions, experienced readers often use conventional cine sequences to accurately identify the myocardium borders.
Purpose: To develop a deep learning model for combining LGE and cine images to improve the robustness and accuracy of LGE scar quantification.
Study type: Retrospective.
Population: A total of 191 hypertrophic cardiomyopathy patients: 1) 162 patients from two sites randomly split into training (50%; 81 patients), validation (25%, 40 patients), and testing (25%; 41 patients); and 2) an external testing dataset (29 patients) from a third site.
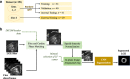
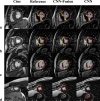
Field strength/sequence: 1.5T, inversion-recovery segmented gradient-echo LGE and balanced steady-state free-precession cine sequences ASSESSMENT: Two convolutional neural networks (CNN) were trained for myocardium and scar segmentation, one with and one without LGE-Cine fusion. For CNN with fusion, the input was two aligned LGE and cine images at matched cardiac phase and anatomical location. For CNN without fusion, only LGE images were used as input. Manual segmentation of the datasets was used as reference standard.
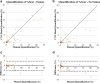
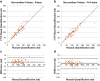
Statistical tests: Manual and CNN-based quantifications of LGE scar burden and of myocardial volume were assessed using Pearson linear correlation coefficients (r) and Bland-Altman analysis.
Results: Both CNN models showed strong agreement with manual quantification of LGE scar burden and myocardium volume. CNN with LGE-Cine fusion was more robust than CNN without LGE-Cine fusion, allowing for successful segmentation of significantly more slices (603 [95%] vs. 562 (89%) of 635 slices; P < 0.001). Also, CNN with LGE-Cine fusion showed better agreement with manual quantification of LGE scar burden than CNN without LGE-Cine fusion (%ScarLGE-cine = 0.82 × %Scarmanual , r = 0.84 vs. %ScarLGE = 0.47 × %Scarmanual , r = 0.81) and myocardium volume (VolumeLGE-cine = 1.03 × Volumemanual , r = 0.96 vs. VolumeLGE = 0.91 × Volumemanual , r = 0.91).
Data conclusion: CNN based LGE-Cine fusion can improve the robustness and accuracy of automated scar quantification.
Level of evidence: 3 TECHNICAL EFFICACY: 1.
Keywords: Deep learning; image fusion; image segmentation; late gadolinium enhancement; myocardial scar; scar quantification.
© 2021 The Authors. Journal of Magnetic Resonance Imaging published by Wiley Periodicals LLC. on behalf of International Society for Magnetic Resonance in Medicine.
Figures
References
-
- Di Marco A, Anguera I, Schmitt M, et al. Late gadolinium enhancement and the risk for ventricular arrhythmias or sudden death in dilated cardiomyopathy. JACC Heart Fail 2017;5(1):28‐38. - PubMed
-
- Disertori M, Rigoni M, Pace N, et al. Myocardial fibrosis assessment by LGE is a powerful predictor of ventricular tachyarrhythmias in ischemic and nonischemic LV dysfunction: A meta‐analysis. JACC Cardiovasc Imaging 2016;9(9):1046‐1055. - PubMed
-
- Chan RH, Maron BJ, Olivotto I, et al. Prognostic value of quantitative contrast‐enhanced cardiovascular magnetic resonance for the evaluation of sudden death risk in patients with hypertrophic cardiomyopathy. Circulation 2014;130(6):484‐495. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

