In silico prediction of SARS-CoV-2 main protease and polymerase inhibitors: 3D-Pharmacophore modelling
- PMID: 33599180
- PMCID: PMC7898304
- DOI: 10.1080/07391102.2021.1886991
In silico prediction of SARS-CoV-2 main protease and polymerase inhibitors: 3D-Pharmacophore modelling
Abstract
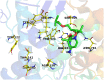
The outbreak of the second severe acute respiratory syndrome coronavirus (SARS-CoV-2) known as COVID-19 has caused global concern. No effective vaccine or treatment to control the virus has been approved yet. Social distancing and precautionary protocols are still the only way to prevent person-to-person transmission. We hope to identify anti-COVID-19 activity of the existing drugs to overcome this pandemic as soon as possible. The present study used HEX and AutoDock Vina softwares to predict the affinity of about 100 medicinal structures toward the active site of 3-chymotrypsin-like protease (3Clpro) and RNA-dependent RNA polymerase (RdRp), separately. Afterwards, MOE software and the pharmacophore-derived query methodology were employed to determine the pharmacophore model of their inhibitors. Tegobuvir (19) and compound 45 showed the best binding affinity toward RdRp and 3Clpro of SARS-CoV-2 in silico, respectively. Tegobuvir -previously applied for hepatitis C virus- formed highly stable complex with uncommon binding pocket of RdRp (E total: -707.91 Kcal/mol) in silico. In addition to compound 45, tipranavir (28) and atazanavir (26) as FDA-approved HIV protease inhibitors were tightly interacted with the active site of SARS-CoV-2 main protease as well. Based on pharmacophore modelling, a good structural pattern for potent candidates against SARS-CoV-2 main enzymes is suggested. Re-tasking or taking inspiration from the structures of tegobuvir and tipranavir can be a proper approach toward coping with the COVID-19 in the shortest possible time and at the lowest cost.Communicated by Ramaswamy H. Sarma.
Keywords: RdRp; SARS-CoV-2; docking study; pharmacophore modelling; protease 3Clpro.
Conflict of interest statement
The authors declare no conflict of interest, financial or otherwise.
Figures








References
-
- Cao, B., Wang, Y., Wen, D., Liu, W., Wang, J., Fan, G., Ruan, L., Song, B., Cai, Y., Wei, M., Li, X., Xia, J., Chen, N., Xiang, J., Yu, T., Bai, T., Xie, X., Zhang, L., Li, C., … Wang, C. (2020). A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe COVID-19. The New England Journal of Medicine, 382(19), 1787–1799. 10.1056/NEJMoa2001282 - DOI - PMC - PubMed
-
- Chan, J. F. W., Kok, K. H., Zhu, Z., Chu, H., To, K. K. W., Yuan, S., & Yuen, K. Y. (2020). Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerging Microbes & Infections, 9(1), 221–236. 10.1080/22221751.2020.1719902 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
