Extreme genetic diversity in the type VII secretion system of Listeria monocytogenes suggests a role in bacterial antagonism
- PMID: 33599605
- PMCID: PMC7613182
- DOI: 10.1099/mic.0.001034
Extreme genetic diversity in the type VII secretion system of Listeria monocytogenes suggests a role in bacterial antagonism
Abstract
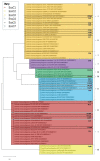
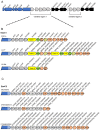
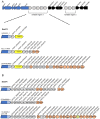
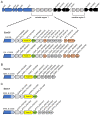
The type VII protein secretion system (T7SS) has been characterized in members of the phyla Actinobacteria and Firmicutes. In mycobacteria the T7SS is intimately linked with pathogenesis and intracellular survival, while in Firmicutes there is mounting evidence that the system plays a key role in interbacterial competition. A conserved membrane-bound ATPase protein, termed EssC in Staphylococcus aureus, is a critical component of the T7SS and is the primary receptor for substrate proteins. Genetic diversity in the essC gene of S. aureus has previously been reported, resulting in four protein variants that are linked to specific subsets of substrates. Here we have analysed the genetic diversity of the T7SS-encoding genes and substrate proteins across Listeria monocytogenes genome sequences. We find that there are seven EssC variants across the species that differ in their C-terminal region; each variant is correlated with a distinct subset of genes for likely substrate and accessory proteins. EssC1 is most common and is exclusively linked with polymorphic toxins harbouring a YeeF domain, whereas EssC5, EssC6 and EssC7 variants all code for an LXG domain protein adjacent to essC. Some essC1 variant strains encode an additional, truncated essC at their T7 gene cluster. The truncated EssC, comprising only the C-terminal half of the protein, matches the sequence of either EssC2, EssC3 or EssC4. In each case the truncated gene directly precedes a cluster of substrate/accessory protein genes acquired from the corresponding strain. Across L. monocytogenes strains we identified 40 LXG domain proteins, most of which are encoded at conserved genomic loci. These loci also harbour genes encoding immunity proteins and sometimes additional toxin fragments. Collectively our findings strongly suggest that the T7SS plays an important role in bacterial antagonism in this species.
Keywords: Listeria monocytogenes; bacterial antagonism; immunity protein; toxin; type VII secretion.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
A novel variant of the Listeria monocytogenes type VII secretion system EssC component is associated with an Rhs toxin.Microb Genom. 2023 Jun;9(6):mgen001036. doi: 10.1099/mgen.0.001036. Microb Genom. 2023. PMID: 37278699 Free PMC article.
-
EssC is a specificity determinant for Staphylococcus aureus type VII secretion.Microbiology (Reading). 2018 May;164(5):816-820. doi: 10.1099/mic.0.000650. Epub 2018 Apr 5. Microbiology (Reading). 2018. PMID: 29620499 Free PMC article.
-
The Type VII Secretion System of Staphylococcus.Annu Rev Microbiol. 2021 Oct 8;75:471-494. doi: 10.1146/annurev-micro-012721-123600. Epub 2021 Aug 3. Annu Rev Microbiol. 2021. PMID: 34343022
-
The ATPases of the mycobacterial type VII secretion system: Structural and mechanistic insights into secretion.Prog Biophys Mol Biol. 2020 May;152:25-34. doi: 10.1016/j.pbiomolbio.2019.11.008. Epub 2019 Nov 22. Prog Biophys Mol Biol. 2020. PMID: 31765647 Review.
-
The role of proteinaceous toxins secreted by Staphylococcus aureus in interbacterial competition.FEMS Microbes. 2024 Feb 28;5:xtae006. doi: 10.1093/femsmc/xtae006. eCollection 2024. FEMS Microbes. 2024. PMID: 38495077 Free PMC article. Review.
Cited by
-
Heterogeneity of the group B streptococcal type VII secretion system and influence on colonization of the female genital tract.Mol Microbiol. 2023 Aug;120(2):258-275. doi: 10.1111/mmi.15115. Epub 2023 Jun 26. Mol Microbiol. 2023. PMID: 37357823 Free PMC article.
-
A type VII secretion system in Group B Streptococcus mediates cytotoxicity and virulence.PLoS Pathog. 2021 Dec 6;17(12):e1010121. doi: 10.1371/journal.ppat.1010121. eCollection 2021 Dec. PLoS Pathog. 2021. PMID: 34871327 Free PMC article.
-
A novel variant of the Listeria monocytogenes type VII secretion system EssC component is associated with an Rhs toxin.Microb Genom. 2023 Jun;9(6):mgen001036. doi: 10.1099/mgen.0.001036. Microb Genom. 2023. PMID: 37278699 Free PMC article.
-
Dual Targeting Factors Are Required for LXG Toxin Export by the Bacterial Type VIIb Secretion System.mBio. 2022 Oct 26;13(5):e0213722. doi: 10.1128/mbio.02137-22. Epub 2022 Aug 29. mBio. 2022. PMID: 36036513 Free PMC article.
-
Heterogeneity of the group B streptococcal type VII secretion system and influence on colonization of the female genital tract.bioRxiv [Preprint]. 2023 Jan 25:2023.01.25.525443. doi: 10.1101/2023.01.25.525443. bioRxiv. 2023. Update in: Mol Microbiol. 2023 Aug;120(2):258-275. doi: 10.1111/mmi.15115. PMID: 36747681 Free PMC article. Updated. Preprint.
References
-
- Costa TR, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, et al. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol. 2015;13:343–359. - PubMed
-
- Bunduc CM, Bitter W, Houben ENG. Structure and Function of the Mycobacterial Type VII Secretion Systems. Annu Rev Microbiol. 2020;74:315–335. - PubMed
-
- Klein TA, Ahmad S, Whitney JC. Contact-Dependent Interbacterial Antagonism Mediated by Protein Secretion Machines. Trends Microbiol. 2020;28:387–400. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

