Effective approaches to discover new microbial metabolites in a large strain library
- PMID: 33599744
- PMCID: PMC9113118
- DOI: 10.1093/jimb/kuab017
Effective approaches to discover new microbial metabolites in a large strain library
Abstract
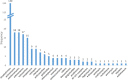
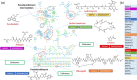
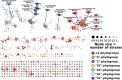
Natural products have provided many molecules to treat and prevent illnesses in humans, animals and plants. While only a small fraction of the existing microbial diversity has been explored for bioactive metabolites, tens of thousands of molecules have been reported in the literature over the past 80 years. Thus, the main challenge in microbial metabolite screening is to avoid the re-discovery of known metabolites in a cost-effective manner. In this perspective, we report and discuss different approaches used in our laboratory over the past few years, ranging from bioactivity-based screening to looking for metabolic rarity in different datasets to deeply investigating a single Streptomyces strain. Our results show that it is possible to find novel chemistry through a limited screening effort, provided that appropriate selection criteria are in place.
Keywords: Planomonospora; Streptomyces; Actinomycetes; Antibiotics; New RiPP family; Pseudouridimycin.
© The Author(s) 2021. Published by Oxford University Press on behalf of Society of Industrial Microbiology and Biotechnology.
Figures



References
-
- Aoyagi T., Kumagai M., Hazato T., Hamada M., Takeuchi T., Umezawa H. (1975). Pyridindolol, a new β-galactosidase inhibitor produced by actinomycetes. Journal of Antibiotics, 28, 555–557. - PubMed
-
- Arnison P. G., Bibb M. J., Bierbaum G., Bowers A. A., Bugni T. S., Bulaj G., Camarero J. A., Campopiano D. J., Challis G. L., Clardy J., Cotter P. D., Craik D. J., Dawson M., Dittmann E., Donadio S., Dorrestein P. C., Entian K. D., Fischbach M. A., Garavelli J. S., van der Donk W. A. (2013). Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Natural Product Reports, 30, 108–160. - PMC - PubMed
-
- Baltz R. H. (2019). Natural product drug discovery in the genomic era: Realities, conjectures, misconceptions and opportunities. Journal of Industrial Microbiology & Biotechnology, 46, 281–299. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

