Differential gene expression analysis of palbociclib-resistant TNBC via RNA-seq
- PMID: 33599863
- PMCID: PMC8019424
- DOI: 10.1007/s10549-021-06127-5
Differential gene expression analysis of palbociclib-resistant TNBC via RNA-seq
Abstract
Purpose: The management of triple-negative breast cancer (TNBC) remains a significant clinical challenge due to the lack of effective targeted therapies. Inhibitors of the cyclin-dependent kinases 4 and 6 (CDK4/6) are emerging as promising therapeutic agents against TNBC; however, cells can rapidly acquire resistance through multiple mechanisms that are yet to be identified. Therefore, determining the mechanisms underlying resistance to CDK4/6 inhibition is crucial to develop combination therapies that can extend the efficacy of the CDK4/6 inhibitors or delay resistance. This study aims to identify differentially expressed genes (DEG) associated with acquired resistance to palbociclib in ER- breast cancer cells.
Methods: We performed next-generation transcriptomic sequencing (RNA-seq) and pathway analysis in ER- MDA-MB-231 palbociclib-sensitive (231/pS) and palbociclib-resistant (231/pR) cells.
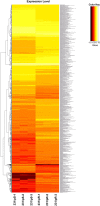
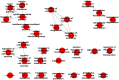
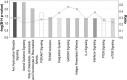
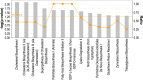
Results: We identified 2247 up-regulated and 1427 down-regulated transcripts in 231/pR compared to 231/pS cells. DEGs were subjected to functional analysis using Gene Ontology (GO) and the KEGG database which identified many transduction pathways associated with breast cancer, including the PI3K/AKT, PTEN and mTOR pathways. Additionally, Ingenuity Pathway Analysis (IPA) revealed that resistance to palbociclib is closely associated with altered cholesterol and fatty acid biosynthesis suggesting that resistance to palbociclib may be dependent on lipid metabolic reprograming.
Conclusion: This study provides evidence that lipid metabolism is altered in TNBC with acquired resistance to palbociclib. Further studies are needed to determine if the observed lipid metabolic rewiring can be exploited to overcome therapy resistance in TNBC.
Keywords: CDK4/6 inhibitors; Metabolism reprogramming; Palbociclib; TNBC; Therapy resistance.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Gupta GK, Collier AL, Lee D, Hoefer RA, Zheleva V, Siewertsz van Reesema LL, Tang-Tan AM, Guye ML, Chang DZ, Winston JS, et al. Perspectives on triple-negative breast cancer: current treatment strategies, unmet needs, and potential targets for future therapies. Cancers (Basel) 2020 doi: 10.3390/cancers12092392. - DOI - PMC - PubMed
-
- Surveillance E, End Results (SEER) Program (www.seer.cancer.gov) SEER*Stat Database: Incidence-SEER Research Data, 9 Registries, Nov 2019 Sub (1975–2017)-Linked To County Attributes-Time Dependent (1990–2017) Income/Rurality, 1969–2017 Counties, National Cancer Institute, DCCPS, Surveillance Research Program, released April 2020, based on the November 2019 submission
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

