A distinct repertoire of cancer-associated fibroblasts is enriched in cribriform prostate cancer
- PMID: 33600062
- PMCID: PMC8073007
- DOI: 10.1002/cjp2.205
A distinct repertoire of cancer-associated fibroblasts is enriched in cribriform prostate cancer
Abstract
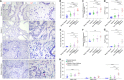
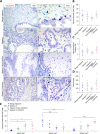
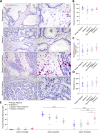
Outcomes for men with localized prostate cancer vary widely, with some men effectively managed without treatment on active surveillance, while other men rapidly progress to metastatic disease despite curative-intent therapies. One of the strongest prognostic indicators of outcome is grade groups based on the Gleason grading system. Gleason grade 4 prostate cancer with cribriform morphology is associated with adverse outcomes and can be utilized clinically to improve risk stratification. The underpinnings of disease aggressiveness associated with cribriform architecture are not fully understood. Most studies have focused on genetic and molecular alterations in cribriform tumor cells; however, less is known about the tumor microenvironment in cribriform prostate cancer. Cancer-associated fibroblasts (CAFs) are a heterogeneous population of fibroblasts in the tumor microenvironment that impact cancer aggressiveness. The overall goal of this study was to determine if cribriform prostate cancers are associated with a unique repertoire of CAFs. Radical prostatectomy whole-tissue sections were analyzed for the expression of fibroblast markers (ASPN in combination with FAP, THY1, ENG, NT5E, TNC, and PDGFRβ) in stroma adjacent to benign glands and in Gleason grade 3, Gleason grade 4 cribriform, and Gleason grade 4 noncribriform prostate cancer by RNAscope®. Halo® Software was used to quantify percent positive stromal cells and expression per positive cell. The fibroblast subtypes enriched in prostate cancer were highly heterogeneous. Both overlapping and distinct populations of low abundant fibroblast subtypes in benign prostate stroma were enriched in Gleason grade 4 prostate cancer with cribriform morphology compared to Gleason grade 4 prostate cancer with noncribriform morphology and Gleason grade 3 prostate cancer. In addition, gene expression was distinctly altered in CAF subtypes adjacent to cribriform prostate cancer. Overall, these studies suggest that cribriform prostate cancer has a unique tumor microenvironment that may distinguish it from other Gleason grade 4 morphologies and lower Gleason grades.
Keywords: ASPN; ENG; FAP; NT5E; PDGFRβ; RNAscope; THY1; cancer; cancer-associated fibroblasts; cribriform; prostate; stroma; tumor microenvironment.
© 2021 The Authors. The Journal of Pathology: Clinical Research published by The Pathological Society of Great Britain and Ireland & John Wiley & Sons, Ltd.
Figures



References
-
- Epstein JI, Amin MB, Reuter VE, et al. Contemporary Gleason grading of prostatic carcinoma: an update with discussion on practical issues to implement the 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. Am J Surg Pathol 2017; 41: e1–e7. - PubMed
-
- Epstein JI. Prostate cancer grading: a decade after the 2005 modified system. Mod Pathol 2018; 31: S47–S63. - PubMed
-
- Gleason DF. Classification of prostatic carcinomas. Cancer Chemother Rep 1966; 50: 125–128. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

