A high-throughput fluorescence lifetime-based assay to detect binding of myosin-binding protein C to F-actin
- PMID: 33600558
- PMCID: PMC7898471
- DOI: 10.1085/jgp.202012707
A high-throughput fluorescence lifetime-based assay to detect binding of myosin-binding protein C to F-actin
Abstract
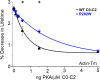
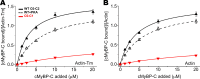
Binding properties of actin-binding proteins are typically evaluated by cosedimentation assays. However, this method is time-consuming, involves multiple steps, and has a limited throughput. These shortcomings preclude its use in screening for drugs that modulate actin-binding proteins relevant to human disease. To develop a simple, quantitative, and scalable F-actin-binding assay, we attached fluorescent probes to actin's Cys-374 and assessed changes in fluorescence lifetime upon binding to the N-terminal region (domains C0-C2) of human cardiac myosin-binding protein C (cMyBP-C). The lifetime of all five probes tested decreased upon incubation with cMyBP-C C0-C2, as measured by time-resolved fluorescence (TR-F), with IAEDANS being the most sensitive probe that yielded the smallest errors. The TR-F assay was compared with cosedimentation to evaluate in vitro changes in binding to actin and actin-tropomyosin arising from cMyBP-C mutations associated with hypertrophic cardiomyopathy (HCM) and tropomyosin binding. Lifetime changes of labeled actin with added C0-C2 were consistent with cosedimentation results. The HCM mutation L352P was confirmed to enhance actin binding, whereas PKA phosphorylation reduced binding. The HCM mutation R282W, predicted to disrupt a PKA recognition sequence, led to deficits in C0-C2 phosphorylation and altered binding. Lastly, C0-C2 binding was found to be enhanced by tropomyosin and binding capacity to be altered by mutations in a tropomyosin-binding region. These findings suggest that the TR-F assay is suitable for rapidly and accurately determining quantitative binding and for screening physiological conditions and compounds that affect cMyBP-C binding to F-actin for therapeutic discovery.
© 2021 Bunch et al.
Figures

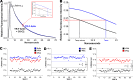


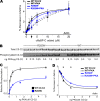


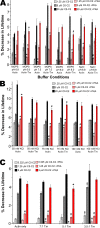
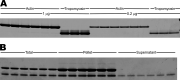
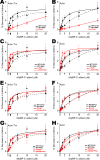


References
-
- Bahrudin, U., Morisaki H., Morisaki T., Ninomiya H., Higaki K., Nanba E., Igawa O., Takashima S., Mizuta E., Miake J., et al. 2008. Ubiquitin-proteasome system impairment caused by a missense cardiac myosin-binding protein C mutation and associated with cardiac dysfunction in hypertrophic cardiomyopathy. J. Mol. Biol. 384:896–907. 10.1016/j.jmb.2008.09.070 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

