Regulation of GFAP Expression
- PMID: 33601918
- PMCID: PMC7897836
- DOI: 10.1177/1759091420981206
Regulation of GFAP Expression
Abstract
Expression of the GFAP gene has attracted considerable attention because its onset is a marker for astrocyte development, its upregulation is a marker for reactive gliosis, and its predominance in astrocytes provides a tool for their genetic manipulation. The literature on GFAP regulation is voluminous, as almost any perturbation of development or homeostasis in the CNS will lead to changes in its expression. In this review, we limit our discussion to mechanisms proposed to regulate GFAP synthesis through a direct interaction with its gene or mRNA. Strengths and weaknesses of the supportive experimental findings are described, and suggestions made for additional studies. This review covers 15 transcription factors, DNA and histone methylation, and microRNAs. The complexity involved in regulating the expression of this intermediate filament protein suggests that GFAP function may vary among both astrocyte subtypes and other GFAP-expressing cells, as well as during development and in response to perturbations.
Keywords: GFAP; astrocyte; gene expression; gene structure; gliogenesis; regulation of transcription and translation.
Conflict of interest statement
Figures
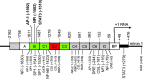

Similar articles
-
AP-1 and the injury response of the GFAP gene.J Neurosci Res. 2019 Feb;97(2):149-161. doi: 10.1002/jnr.24338. Epub 2018 Oct 22. J Neurosci Res. 2019. PMID: 30345544 Free PMC article.
-
GFAP promoter directs astrocyte-specific expression in transgenic mice.J Neurosci. 1994 Mar;14(3 Pt 1):1030-7. doi: 10.1523/JNEUROSCI.14-03-01030.1994. J Neurosci. 1994. PMID: 8120611 Free PMC article.
-
GFAP promoter elements required for region-specific and astrocyte-specific expression.Glia. 2008 Apr;56(5):481-93. doi: 10.1002/glia.20622. Glia. 2008. PMID: 18240313
-
Neurochemical regulation of the expression and function of glial fibrillary acidic protein in astrocytes.Glia. 2020 May;68(5):878-897. doi: 10.1002/glia.23734. Epub 2019 Oct 18. Glia. 2020. PMID: 31626364 Review.
-
Glial fibrillary acidic protein: from intermediate filament assembly and gliosis to neurobiomarker.Trends Neurosci. 2015 Jun;38(6):364-74. doi: 10.1016/j.tins.2015.04.003. Epub 2015 May 11. Trends Neurosci. 2015. PMID: 25975510 Free PMC article. Review.
Cited by
-
Overexpression of pathogenic tau in astrocytes causes a reduction in AQP4 and GLT1, an immunosuppressed phenotype and unique transcriptional responses to repetitive mild TBI without appreciable changes in tauopathy.J Neuroinflammation. 2024 May 15;21(1):130. doi: 10.1186/s12974-024-03117-4. J Neuroinflammation. 2024. PMID: 38750510 Free PMC article.
-
Extracellular Vesicles Derived from Young Neural Cultures Attenuate Astrocytic Reactivity In Vitro.Int J Mol Sci. 2022 Jan 25;23(3):1371. doi: 10.3390/ijms23031371. Int J Mol Sci. 2022. PMID: 35163295 Free PMC article.
-
Delivery of miR-15b-5p via magnetic nanoparticle-enhanced bone marrow mesenchymal stem cell-derived extracellular vesicles mitigates diabetic osteoporosis by targeting GFAP.Cell Biol Toxicol. 2024 Jul 5;40(1):52. doi: 10.1007/s10565-024-09877-2. Cell Biol Toxicol. 2024. PMID: 38967699 Free PMC article.
-
The Biflavonoid Agathisflavone Regulates Microglial and Astrocytic Inflammatory Profiles via Glucocorticoid Receptor.Molecules. 2025 Feb 22;30(5):1014. doi: 10.3390/molecules30051014. Molecules. 2025. PMID: 40076239 Free PMC article.
-
The Comparison of the Selected Parameters of Brain Injury and Interleukins in the CSF in Patients Diagnosed De Novo with RRMS Compared to the Control Group.Diagnostics (Basel). 2023 Nov 13;13(22):3436. doi: 10.3390/diagnostics13223436. Diagnostics (Basel). 2023. PMID: 37998571 Free PMC article.
References
-
- Abidin S. Z., Fam S. Z., Chong C. E., Abdullah S., Cheah P. S., Nordin N., Ling K. H. (2019. ) miR-3099 promotes neurogenesis and inhibits astrogliogenesis during murine neural development. Gene, 697, 201–212. - PubMed
-
- Asano H., Aonuma M., Sanosaka T., Kohyama J., Namihira M., Nakashima K. (2009) Astrocyte differentiation of neural precursor cells is enhanced by retinoic acid through a change in epigenetic modification. Stem Cells, 27, 2744–2752. - PubMed
-
- Bachetti T., Di Zanni E., Lantieri F., Caroli F., Regis S., Filocamo M., Rainero I., Gallone S., Cilia R., Romano S., Savoiardo M., Pareyson D., Biancheri R., Ravazzolo R., Ceccherini I. (2010) A novel polymorphic AP-1 binding element of the GFAP promoter is associated with different allelic transcriptional activities. Ann Hum Genet, 74, 506–515. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
