Addressing uncertainty in genome-scale metabolic model reconstruction and analysis
- PMID: 33602294
- PMCID: PMC7890832
- DOI: 10.1186/s13059-021-02289-z
Addressing uncertainty in genome-scale metabolic model reconstruction and analysis
Abstract
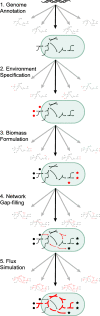
The reconstruction and analysis of genome-scale metabolic models constitutes a powerful systems biology approach, with applications ranging from basic understanding of genotype-phenotype mapping to solving biomedical and environmental problems. However, the biological insight obtained from these models is limited by multiple heterogeneous sources of uncertainty, which are often difficult to quantify. Here we review the major sources of uncertainty and survey existing approaches developed for representing and addressing them. A unified formal characterization of these uncertainties through probabilistic approaches and ensemble modeling will facilitate convergence towards consistent reconstruction pipelines, improved data integration algorithms, and more accurate assessment of predictive capacity.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

