The DUF1013 protein TrcR tracks with RNA polymerase to control the bacterial cell cycle and protect against antibiotics
- PMID: 33602809
- PMCID: PMC7923619
- DOI: 10.1073/pnas.2010357118
The DUF1013 protein TrcR tracks with RNA polymerase to control the bacterial cell cycle and protect against antibiotics
Abstract
How DNA-dependent RNA polymerase (RNAP) acts on bacterial cell cycle progression during transcription elongation is poorly investigated. A forward genetic selection for Caulobacter crescentus cell cycle mutants unearthed the uncharacterized DUF1013 protein (TrcR, transcriptional cell cycle regulator). TrcR promotes the accumulation of the essential cell cycle transcriptional activator CtrA in late S-phase but also affects transcription at a global level to protect cells from the quinolone antibiotic nalidixic acid that induces a multidrug efflux pump and from the RNAP inhibitor rifampicin that blocks transcription elongation. We show that TrcR associates with promoters and coding sequences in vivo in a rifampicin-dependent manner and that it interacts physically and genetically with RNAP. We show that TrcR function and its RNAP-dependent chromatin recruitment are conserved in symbiotic Sinorhizobium sp. and pathogenic Brucella spp Thus, TrcR represents a hitherto unknown antibiotic target and the founding member of the DUF1013 family, an uncharacterized class of transcriptional regulators that track with RNAP during the elongation phase to promote transcription during the cell cycle.
Keywords: Brucella; Caulobacter; RNA polymerase; TrcR; transcription.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures
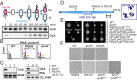
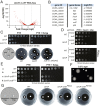




References
-
- Ebright R. H., RNA polymerase: Structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J. Mol. Biol. 304, 687–698 (2000). - PubMed
-
- Darst S. A., Bacterial RNA polymerase. Curr. Opin. Struct. Biol. 11, 155–162 (2001). - PubMed
-
- Browning D. F., Busby S. J., The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2, 57–65 (2004). - PubMed
-
- Browning D. F., Busby S. J., Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. 14, 638–650 (2016). - PubMed
-
- Perederina A., et al. ., Regulation through the secondary channel–structural framework for ppGpp-DksA synergism during transcription. Cell 118, 297–309 (2004). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

