Sophisticated viral quasispecies with a genotype-related pattern of mutations in the hepatitis B X gene of HBeAg-ve chronically infected patients
- PMID: 33603102
- PMCID: PMC7892877
- DOI: 10.1038/s41598-021-83762-4
Sophisticated viral quasispecies with a genotype-related pattern of mutations in the hepatitis B X gene of HBeAg-ve chronically infected patients
Abstract
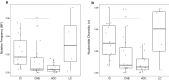
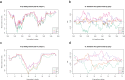
Patients with HBeAg-negative chronic infection (CI) have not been extensively studied because of low viremia. The HBx protein, encoded by HBX, has a key role in viral replication. Here, we analyzed the viral quasispecies at the 5' end of HBX in CI patients and compared it with that of patients in other clinical stages. Fifty-eight HBeAg-negative patients were included: 16 CI, 19 chronic hepatitis B, 16 hepatocellular carcinoma and 6 liver cirrhosis. Quasispecies complexity and conservation were determined in the region between nucleotides 1255 and 1611. Amino acid changes detected were tested in vitro. CI patients showed higher complexity in terms of mutation frequency and nucleotide diversity and higher quasispecies conservation (p < 0.05). A genotype D-specific pattern of mutations (A12S/P33S/P46S/T36D-G) was identified in CI (median frequency, 81.7%), which determined a reduction in HBV DNA release of up to 1.5 log in vitro. CI patients showed a more complex and conserved viral quasispecies than the other groups. The genotype-specific pattern of mutations could partially explain the low viremia observed in these patients.
Conflict of interest statement
Josep Gregori is an employee of Roche Diagnostics, SL. No competing interests are reported for the other authors.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

