Potential of spectroscopic analyses for non-destructive estimation of tea quality-related metabolites in fresh new leaves
- PMID: 33603126
- PMCID: PMC7892543
- DOI: 10.1038/s41598-021-83847-0
Potential of spectroscopic analyses for non-destructive estimation of tea quality-related metabolites in fresh new leaves
Abstract
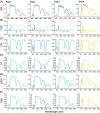
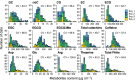
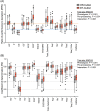
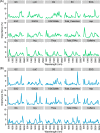
Spectroscopic sensing provides physical and chemical information in a non-destructive and rapid manner. To develop non-destructive estimation methods of tea quality-related metabolites in fresh leaves, we estimated the contents of free amino acids, catechins, and caffeine in fresh tea leaves using visible to short-wave infrared hyperspectral reflectance data and machine learning algorithms. We acquired these data from approximately 200 new leaves with various status and then constructed the regression model in the combination of six spectral patterns with pre-processing and five algorithms. In most phenotypes, the combination of de-trending pre-processing and Cubist algorithms was robustly selected as the best combination in each round over 100 repetitions that were evaluated based on the ratio of performance to deviation (RPD) values. The mean RPD values were ranged from 1.1 to 2.7 and most of them were above the acceptable or accurate threshold (RPD = 1.4 or 2.0, respectively). Data-based sensitivity analysis identified the important hyperspectral regions around 1500 and 2000 nm. Present spectroscopic approaches indicate that most tea quality-related metabolites can be estimated non-destructively, and pre-processing techniques help to improve its accuracy.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

