Robust decomposition of cell type mixtures in spatial transcriptomics
- PMID: 33603203
- PMCID: PMC8606190
- DOI: 10.1038/s41587-021-00830-w
Robust decomposition of cell type mixtures in spatial transcriptomics
Abstract
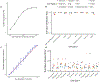
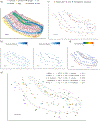
A limitation of spatial transcriptomics technologies is that individual measurements may contain contributions from multiple cells, hindering the discovery of cell-type-specific spatial patterns of localization and expression. Here, we develop robust cell type decomposition (RCTD), a computational method that leverages cell type profiles learned from single-cell RNA-seq to decompose cell type mixtures while correcting for differences across sequencing technologies. We demonstrate the ability of RCTD to detect mixtures and identify cell types on simulated datasets. Furthermore, RCTD accurately reproduces known cell type and subtype localization patterns in Slide-seq and Visium datasets of the mouse brain. Finally, we show how RCTD's recovery of cell type localization enables the discovery of genes within a cell type whose expression depends on spatial environment. Spatial mapping of cell types with RCTD enables the spatial components of cellular identity to be defined, uncovering new principles of cellular organization in biological tissue. RCTD is publicly available as an open-source R package at https://github.com/dmcable/RCTD .
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Conflict of Interest Statement
The authors declare no conflict of interest.
Figures




References
-
- Stickels RR et al. Sensitive spatial genome wide expression profiling at cellular resolution. bioRxiv (2020). https://www.biorxiv.org/content/early/2020/03/14/2020.03.12.989806.full.pdf.
-
- 10x Genomics. 10x genomics: Visium spatial gene expression. https://www.10xgenomics.com/solutions/spatial-gene-expression/ (2020).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

