Evaluation of FGFR1 as a diagnostic biomarker for ovarian cancer using TCGA and GEO datasets
- PMID: 33604191
- PMCID: PMC7866899
- DOI: 10.7717/peerj.10817
Evaluation of FGFR1 as a diagnostic biomarker for ovarian cancer using TCGA and GEO datasets
Abstract
Background: Malignant ovarian cancer is associated with the highest mortality of all gynecological tumors. Designing therapeutic targets that are specific to OC tissue is important for optimizing OC therapies. This study aims to identify different expression patterns of genes related to FGFR1 and the usefulness of FGFR1 as diagnostic biomarker for OC.
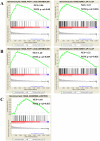
Methods: We collected data from The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO) databases. In the TCGA cohort we analyzed clinical information according to patient characteristics, including age, stage, grade, longest dimension of the tumor and the presence of a residual tumor. GEO data served as a validation set. We obtained data on differentially expressed genes (DEGs) from the two microarray datasets. We then used gene set enrichment analysis (GSEA) to analyze the DEG data in order to identify enriched pathways related to FGFR1.
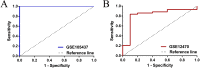
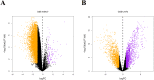
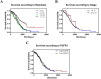
Results: Differential expression analysis revealed that FGFR1 was significantly downregulated in OC specimens. 303 patients were included in the TCGA cohort. The GEO dataset confirmed these findings using information on 75 Asian patients. The GSE105437 and GSE12470 database highlighted the significant diagnostic value of FGFR1 in identifying OC (AUC = 1, p = 0.0009 and AUC = 0.8256, p = 0.0015 respectively).
Conclusions: Our study examined existing TCGA and GEO datasets for novel factors associated with OC and identified FGFR1 as a potential diagnostic factor. Further investigation is warranted to characterize the role played by FGFR1 in OC.
Keywords: FGFR1; Ovarian cancer; RNA-sequencing; Targeted treatment.
©2021 Xiao et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures


Similar articles
-
Identification and Validation of the Diagnostic Characteristic Genes of Ovarian Cancer by Bioinformatics and Machine Learning.Front Genet. 2022 Jun 1;13:858466. doi: 10.3389/fgene.2022.858466. eCollection 2022. Front Genet. 2022. PMID: 35719392 Free PMC article.
-
Multifactor assessment of ovarian cancer reveals immunologically interpretable molecular subtypes with distinct prognoses.Front Immunol. 2023 Dec 7;14:1326018. doi: 10.3389/fimmu.2023.1326018. eCollection 2023. Front Immunol. 2023. PMID: 38143770 Free PMC article.
-
Integrated bioinformatics analysis for the screening of hub genes and therapeutic drugs in ovarian cancer.J Ovarian Res. 2020 Jan 27;13(1):10. doi: 10.1186/s13048-020-0613-2. J Ovarian Res. 2020. PMID: 31987036 Free PMC article.
-
Prospective pathway signaling and prognostic values of MicroRNA-9 in ovarian cancer based on gene expression omnibus (GEO): a bioinformatics analysis.J Ovarian Res. 2021 Feb 9;14(1):29. doi: 10.1186/s13048-021-00779-z. J Ovarian Res. 2021. PMID: 33563317 Free PMC article.
-
Inferring Aberrant Signal Transduction Pathways in Ovarian Cancer from TCGA Data.Cancer Inform. 2014 Oct 13;13(Suppl 1):29-36. doi: 10.4137/CIN.S13881. eCollection 2014. Cancer Inform. 2014. PMID: 25392681 Free PMC article. Review.
Cited by
-
Analysis and identification of oxidative stress-ferroptosis related biomarkers in ischemic stroke.Sci Rep. 2024 Feb 15;14(1):3803. doi: 10.1038/s41598-024-54555-2. Sci Rep. 2024. PMID: 38360841 Free PMC article.
-
ATF4 renders human T-cell acute lymphoblastic leukemia cell resistance to FGFR1 inhibitors through amino acid metabolic reprogramming.Acta Pharmacol Sin. 2023 Nov;44(11):2282-2295. doi: 10.1038/s41401-023-01108-4. Epub 2023 Jun 6. Acta Pharmacol Sin. 2023. PMID: 37280363 Free PMC article.
-
A pan-cancer analysis of the oncogenic role of Keratin 17 (KRT17) in human tumors.Transl Cancer Res. 2021 Oct;10(10):4489-4501. doi: 10.21037/tcr-21-2118. Transl Cancer Res. 2021. PMID: 35116305 Free PMC article.
-
Signatures of tumor-associated macrophages correlate with treatment response in ovarian cancer patients.Aging (Albany NY). 2024 Jan 3;16(1):207-225. doi: 10.18632/aging.205362. Epub 2024 Jan 3. Aging (Albany NY). 2024. PMID: 38175687 Free PMC article.
-
Leucine-rich repeat-containing protein 19 suppresses colorectal cancer by targeting cyclin-dependent kinase 6/E2F1 and remodeling the immune microenvironment.World J Gastroenterol. 2025 Jul 7;31(25):107893. doi: 10.3748/wjg.v31.i25.107893. World J Gastroenterol. 2025. PMID: 40656610 Free PMC article.
References
-
- Beltrame L, Marino DM, Fruscio R, Calura E, Chapman B, Clivio L, Sina F, Mele C, Iatropoulos P, Grassi T, Fotia V, Romualdi C, Martini P, Noris M, Paracchini L, Craparotta I, Petrillo M, Milani R, Perego P, Ravaggi A, Zambelli A, Ronchetti E, D’Incalci M, Marchini S. Profiling cancer gene mutations in longitudinal epithelial ovarian cancer biopsies by targeted next-generation sequencing: a retrospective study. Annals of Oncology Official Journal of the European Society for Medical Oncology. 2015;26:1363–1371. doi: 10.1093/annonc/mdv164. - DOI - PubMed
-
- Bennouna J, Hiret S, Bertaut A, Bouche O, Deplanque G, Borel C, Francois E, Conroy T, Ghiringhelli F, Guetz GDes, Seitz JF, Artru P, Hebbar M, Stanbury T, Denis MG, Adenis A, Borg C. Continuation of bevacizumab vs cetuximab plus chemotherapy after first progression in kras wild-type metastatic colorectal cancer: the UNICANCER PRODIGE18 randomized clinical trial. JAMA Oncology. 2018;5(1):83–90. doi: 10.1001/jamaoncol.2018.4465. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

