Metagenomic Analysis of the Gut Microbiota of Wild Mice, a Newly Identified Reservoir of Campylobacter
- PMID: 33604305
- PMCID: PMC7884769
- DOI: 10.3389/fcimb.2020.596149
Metagenomic Analysis of the Gut Microbiota of Wild Mice, a Newly Identified Reservoir of Campylobacter
Abstract
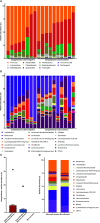
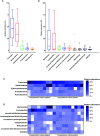
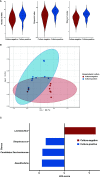
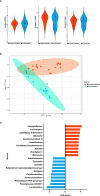
Campylobacter, the most common etiologic agent of zoonotic gastroenteritis in humans, is present in many reservoirs including livestock animals, wildlife, soil, and water. Previously, we reported a novel Campylobacter jejuni strain SCJK02 (MLST ST-8388) from the gut of wild mice (Micromys minutus) using culture-dependent methods. However, due to fastidious growth conditions and the presence of viable but non-culturable Campylobacter spp., it is unclear whether M. minutus is a Campylobacter reservoir. This study aimed to: 1) determine the distribution and proportion of Campylobacter spp. in the gut microbiota of wild mice using culture-independent methods and 2) investigate the gut microbiota of wild mice and the relationship of Campylobacter spp. with other gut microbes. The gut microbiota of 38 wild mice captured from perilla fields in Korea and without any clinical symptoms (18 M. minutus and 20 Mus musculus) were analyzed. Metagenomic analysis showed that 77.8% (14 of 18) of the captured M. minutus harbored Campylobacter spp. (0.24-32.92%) in the gut metagenome, whereas none of the captured M. musculus carried Campylobacter spp. in their guts. Notably, 75% (6 of 8) of M. minutus determined to be Campylobacter-negative using culture-dependent methods showed a high proportion of Campylobacter through metagenome analysis. The results of metagenome analysis and the absence of clinical symptoms suggest that Campylobacter may be a component of the normal gut flora of wild M. minutus. Furthermore, linear discriminant analysis (LDA) showed that Campylobacter was the most enriched genus in the gut microbiota of M. minutus (LDA score, 5.37), whereas Lactobacillus was the most enriched genus in M. musculus (LDA score, -5.96). The differences in the presence of Campylobacter between the two species of wild mice may be attributed to the differential abundance of Campylobacter and Lactobacillus in their respective gut microbiota. In conclusion, the results indicate that wild M. minutus may serve as a potential Campylobacter reservoir. This study presents the first metagenomics analysis of the M. minutus gut microbiota to explore its possible role as an environmental Campylobacter reservoir and provides a basis for future studies using culture-independent methods to determine the role of environmental reservoirs in Campylobacter transmission.
Keywords: Campylobacter; Lactobacillus; Micromys minutus; environmental reservoir; gut microbiota; metagenomics; transmission cycle; wild mouse.
Copyright © 2021 Song, Kim, Guk, Kim, Nam, Suh, Seong and Cho.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
The Wild Mouse (Micromys minutus): Reservoir of a Novel Campylobacter jejuni Strain.Front Microbiol. 2020 Jan 14;10:3066. doi: 10.3389/fmicb.2019.03066. eCollection 2019. Front Microbiol. 2020. PMID: 31993041 Free PMC article.
-
Occurrence and molecular analysis of Campylobacter in wildlife on livestock farms.Vet Microbiol. 2012 Jun 15;157(3-4):369-75. doi: 10.1016/j.vetmic.2011.12.026. Epub 2011 Dec 29. Vet Microbiol. 2012. PMID: 22266157
-
Characterisation by multilocus sequence and porA and flaA typing of Campylobacter jejuni isolated from samples of dog faeces collected in one city in New Zealand.N Z Vet J. 2017 Jul;65(4):209-213. doi: 10.1080/00480169.2017.1311810. Epub 2017 Apr 26. N Z Vet J. 2017. PMID: 28372482
-
Re-thinking the chicken-Campylobacter jejuni interaction: a review.Avian Pathol. 2018 Aug;47(4):352-363. doi: 10.1080/03079457.2018.1475724. Epub 2018 Jun 11. Avian Pathol. 2018. PMID: 29764197 Review.
-
The human gut microbiome, a taxonomic conundrum.Syst Appl Microbiol. 2015 Jun;38(4):276-86. doi: 10.1016/j.syapm.2015.03.004. Epub 2015 Mar 25. Syst Appl Microbiol. 2015. PMID: 25864640 Review.
Cited by
-
Probiogenomic In-Silico Analysis and Safety Assessment of Lactiplantibacillus plantarum DJF10 Strain Isolated from Korean Raw Milk.Int J Mol Sci. 2022 Nov 21;23(22):14494. doi: 10.3390/ijms232214494. Int J Mol Sci. 2022. PMID: 36430971 Free PMC article.
-
From germ-free to wild: modulating microbiome complexity to understand mucosal immunology.Mucosal Immunol. 2022 Jun;15(6):1085-1094. doi: 10.1038/s41385-022-00562-3. Epub 2022 Sep 5. Mucosal Immunol. 2022. PMID: 36065057 Review.
-
Campylobacter in Africa - A specific viewpoint.Eur J Microbiol Immunol (Bp). 2023 Dec 5;13(4):107-124. doi: 10.1556/1886.2023.00043. Print 2023 Dec 21. Eur J Microbiol Immunol (Bp). 2023. PMID: 38051352 Free PMC article. Review.
-
New insights into the microbiota of wild mice.Mamm Genome. 2021 Aug;32(4):311-318. doi: 10.1007/s00335-021-09887-z. Epub 2021 Jul 9. Mamm Genome. 2021. PMID: 34241667 Free PMC article. Review.
-
Characterization of captive and wild 13-lined ground squirrel cecal microbiotas using Illumina-based sequencing.Anim Microbiome. 2022 Jan 3;4(1):1. doi: 10.1186/s42523-021-00154-9. Anim Microbiome. 2022. PMID: 34980290 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

