ELAC2, an Enzyme for tRNA Maturation, Plays a Role in the Cleavage of a Mature tRNA to Produce a tRNA-Derived RNA Fragment During Respiratory Syncytial Virus Infection
- PMID: 33604354
- PMCID: PMC7884774
- DOI: 10.3389/fmolb.2020.609732
ELAC2, an Enzyme for tRNA Maturation, Plays a Role in the Cleavage of a Mature tRNA to Produce a tRNA-Derived RNA Fragment During Respiratory Syncytial Virus Infection
Abstract
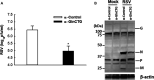
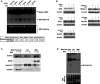
Respiratory syncytial virus (RSV) is the most common cause of lower respiratory tract infection in young children. However, effective treatment against RSV is unavailable. tRNA-derived RNA fragments (tRFs) are a recently discovered family of non-coding RNAs. We made an early observation that RSV infection causes significant induction of tRFs, which are mainly derived from the 5'-end of mature tRNAs (tRF5). However, their functions and biogenesis mechanism are not fully understood. Herein, we identified an enzyme responsible for the induction of a functional tRF5 derived from tRNA-Gln-CTG (tRF5-GlnCTG). We found that tRF5-GlnCTG promotes RSV replication and its induction, assessed by Northern blot and a new qRT-PCR-based method, is regulated by ribonuclease ELAC2. ELAC2-mediated tRF5 induction has never been reported. We also found that ELAC2 is associated with RSV N and NS1 proteins. Given the fact that tRF5-GlnCTG plays a role in RSV replication, the identification of ELAC2 being responsible for tRF5-GlnCTG induction could provide new insights into therapeutic strategy development against RSV infection.
Keywords: ELAC2; RSV; biogenesis; tRF; viral replication.
Copyright © 2021 Choi, Wu, Zhang, Lee, Kim, Lee and Bao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Identification of two novel functional tRNA-derived fragments induced in response to respiratory syncytial virus infection.J Gen Virol. 2017 Jul;98(7):1600-1610. doi: 10.1099/jgv.0.000852. Epub 2017 Jul 15. J Gen Virol. 2017. PMID: 28708049 Free PMC article.
-
Role of Poly(A)-Binding Protein Cytoplasmic 1, a tRNA-Derived RNA Fragment-Bound Protein, in Respiratory Syncytial Virus Infection.Pathogens. 2024 Sep 12;13(9):791. doi: 10.3390/pathogens13090791. Pathogens. 2024. PMID: 39338982 Free PMC article.
-
The Importance of AGO 1 and 4 in Post-Transcriptional Gene Regulatory Function of tRF5-GluCTC, an Respiratory Syncytial Virus-Induced tRNA-Derived RNA Fragment.Int J Mol Sci. 2020 Nov 20;21(22):8766. doi: 10.3390/ijms21228766. Int J Mol Sci. 2020. PMID: 33233493 Free PMC article.
-
tRNA-derived fragments: Mechanisms underlying their regulation of gene expression and potential applications as therapeutic targets in cancers and virus infections.Theranostics. 2021 Jan 1;11(1):461-469. doi: 10.7150/thno.51963. eCollection 2021. Theranostics. 2021. PMID: 33391486 Free PMC article. Review.
-
Non-Coding RNAs and Their Role in Respiratory Syncytial Virus (RSV) and Human Metapneumovirus (hMPV) Infections.Viruses. 2020 Mar 21;12(3):345. doi: 10.3390/v12030345. Viruses. 2020. PMID: 32245206 Free PMC article. Review.
Cited by
-
The role of tRNA-derived small RNAs in aging.BMB Rep. 2023 Feb;56(2):49-55. doi: 10.5483/BMBRep.2022-0199. BMB Rep. 2023. PMID: 36646437 Free PMC article. Review.
-
Parent tRNA Modification Status Determines the Induction of Functional tRNA-Derived RNA by Respiratory Syncytial Virus Infection.Viruses. 2022 Dec 24;15(1):57. doi: 10.3390/v15010057. Viruses. 2022. PMID: 36680097 Free PMC article.
-
The hidden RNA code: implications of the RNA epitranscriptome in the context of viral infections.Front Genet. 2023 Aug 1;14:1245683. doi: 10.3389/fgene.2023.1245683. eCollection 2023. Front Genet. 2023. PMID: 37614818 Free PMC article. Review.
-
tRNA derived fragments:A novel player in gene regulation and applications in cancer.Front Oncol. 2023 Jan 20;13:1063930. doi: 10.3389/fonc.2023.1063930. eCollection 2023. Front Oncol. 2023. PMID: 36761955 Free PMC article. Review.
-
tRNA derived small RNAs-Small players with big roles.Front Genet. 2022 Sep 19;13:997780. doi: 10.3389/fgene.2022.997780. eCollection 2022. Front Genet. 2022. PMID: 36199575 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

